Peter Andorfer, Carolin-Isabel Kahlig, Doris Pakusic, et al. (2024)
Cas-CLOVER-mediated Knockout of STAT1: A Novel Approach to Engineer Packaging HEK-293 Cell Lines Used for rAAV Production
Published in Biotechnology Journal
Abstract
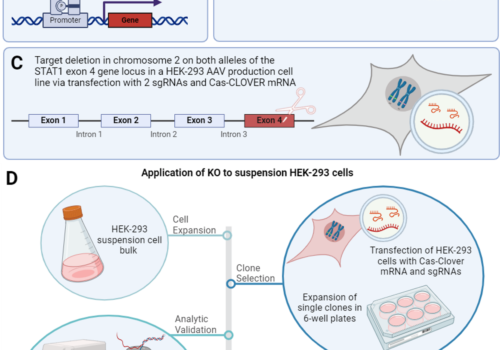
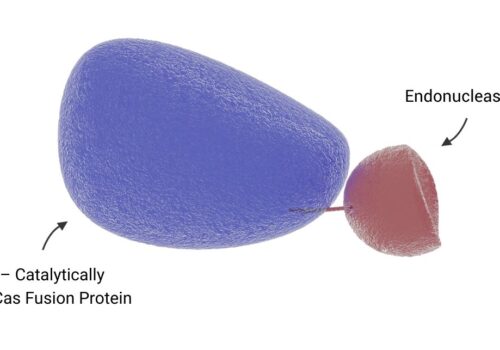
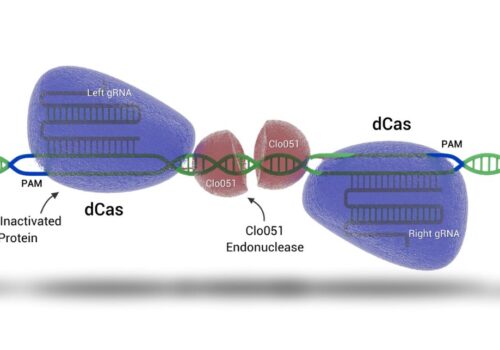
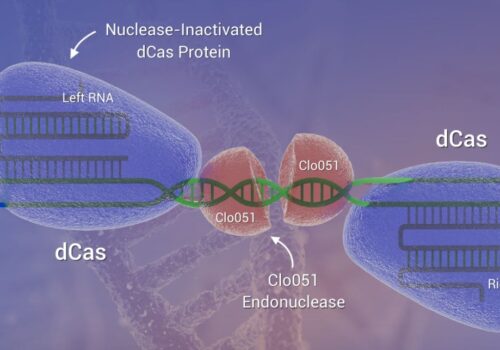
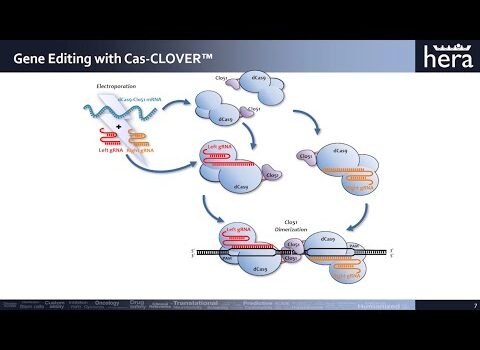
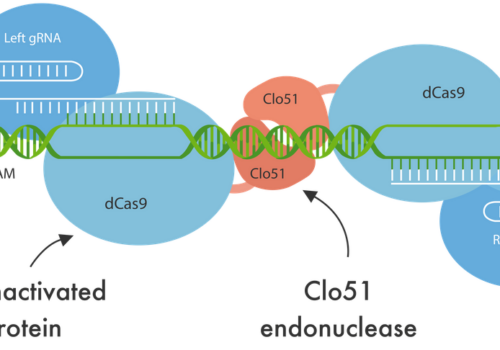
To address the limitations of CRISPR-Cas9, including off-target effects and high licensing costs for commercial use, the Cas-CLOVER system, a dimeric gene-editing tool activated by two guide RNAs, was recently developed. This study evaluates the application of Cas-CLOVER in suspension HEK-293 cells used for recombinant adeno-associated virus (rAAV) production by targeting the STAT1 locus, a critical regulator of cell growth that may influence rAAV production yields.
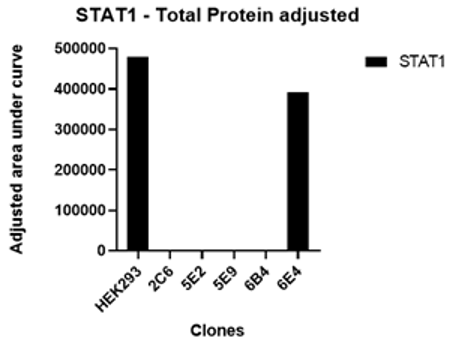
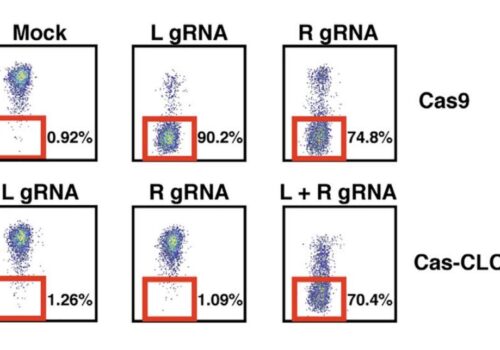
Cas-CLOVER demonstrated high editing efficiency, with mutations detected in all sequencing reads for 12 of the 13 clones analyzed, representing a success rate of 92.3%, consistent with the highest values reported in the literature. All target loci were edited, with no wild-type sequences detected among the edited clones. The mutations primarily consisted of various deletions, with one instance of an insertion, ranging from 3 to 171 base pairs. These larger deletions increased the likelihood of achieving a disruptive knockout, as evidenced by the absence of detectable STAT1 protein via Western blot analysis, regardless of whether the genomic alterations resulted in a frameshift with a subsequent stop codon or an in-frame deletion.
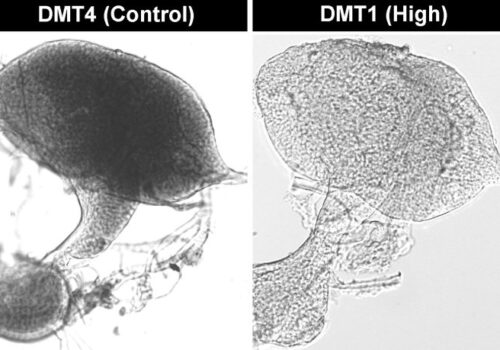
STAT1 is known to play a role in the regulation of cell growth and survival, a finding consistent with the observation of reduced cell numbers in all clones at harvest. Interestingly, only two clones, both triploid, exhibited reduced viability, suggesting that the disruption of three copies of the STAT1 allele had a more severe impact. None of the clones showed an absolute increase in capsid titer, and the vector genome titers were lower in knockout clones. However, STAT1 knockout clones exhibited intracellular capsid and viral replication protein levels up to threefold higher compared to the wild-type control, with significant variation in protein composition. These results highlight the complex regulation of viral proteins and the influence of clonal variation.
In summary, this study demonstrates the applicability of the Cas-CLOVER system in an rAAV-producing HEK-293 cell line. The high editing efficiency and minimal off-target effects make Cas-CLOVER a promising alternative to the standard CRISPR/Cas9 system. Although single STAT1 knockouts did not enhance rAAV yield, the potential for engineering cell lines for improved AAV production remains substantial. Leveraging gene-editing tools like Cas-CLOVER, alongside the genetic diversity of HEK-293 cells and high-throughput technologies, could lead to significant yield improvements.
Peter Andorfer, Carolin-Isabel Kahlig, Doris Pakusic, et al. Cas-CLOVER-mediated knockout of STAT1: A novel approach to engineer packaging HEK-293 cell lines used for rAAV production. Biotechnology Journal.
Additional Resources
More to help guide your gene editing implementation journey