In another blog post, we examined the utilization of Dihydrofolate Reductase (DHFR) inhibitor Methotrexate (MTX) and Glutamine Synthetase (GS) inhibitor Methionine Sulfoximine (MSX) by scientists specializing in cell line development (CLD) to enhance the selection process for cells exhibiting high expression levels. However, these methods require extended durations for selection, and inefficacy results in the inadvertent selection of some low expressing cells. Given that multiple biopharmaceutical product leads must be biomanufactured for preclinical and clinical evaluation prior to the final candidate selection, it is critical that CLD methodologies are optimized for maximum efficiency.
We further examined a study in which researchers determined that the genetic modification of mammalian expression systems may represent a viable solution. Among these systems, Chinese hamster ovary (CHO) cells are predominantly utilized. Initially, the gene editing technology known as Zinc Finger Nuclease (ZFN) was employed to completely inactivate the GS gene, resulting in a more stringent selection process for clones with high production capabilities. However, ZFNs require intense protein engineering and have low gene editing efficiency.
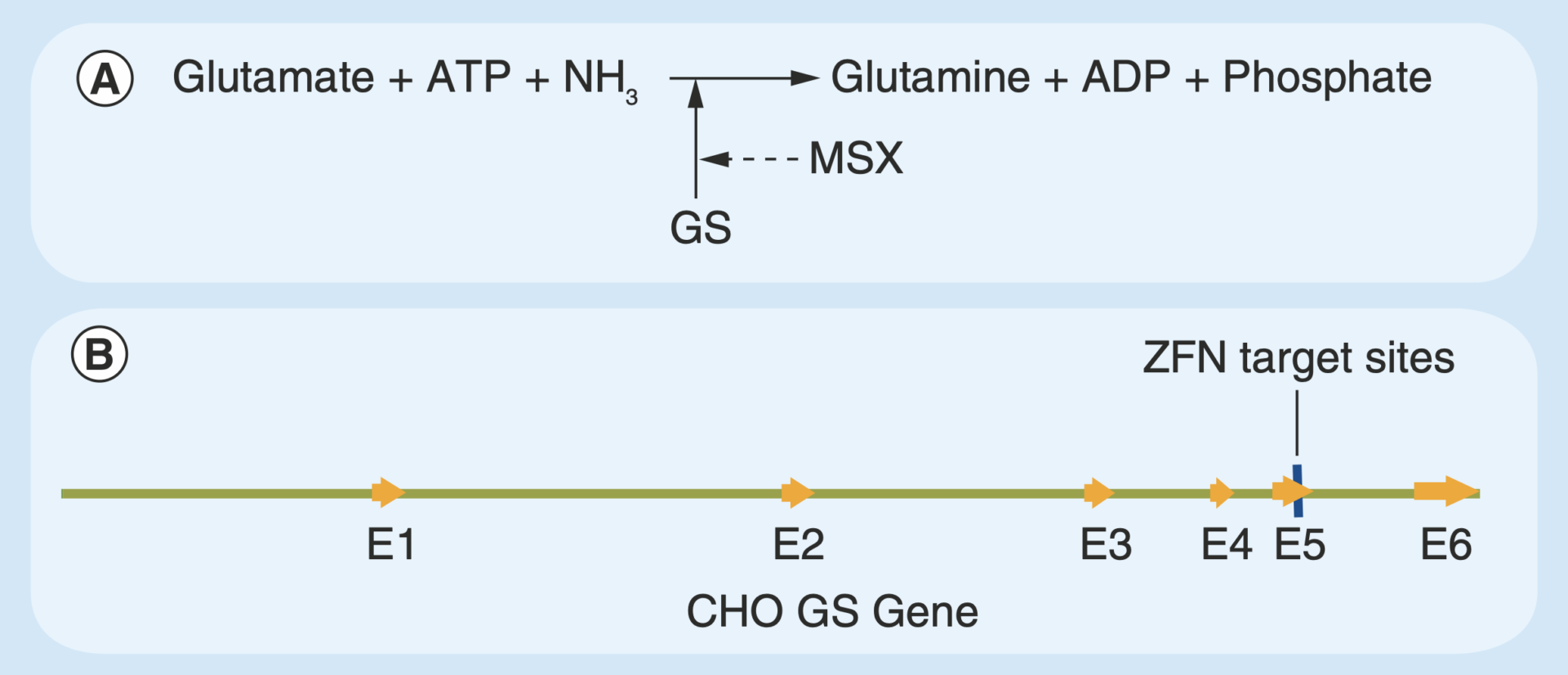
Glutamine synthetase in CHO cells.
(A) GS is crucial in metabolizing nitrogen; it catalyzes glutamate and ammonia to condense glutamine and can be inhibited by MSX; (B) Gene structure of CHO GS locus and ZFN target loci.
Role Of Gene Editing To Enhance CHO Expression Systems
CHO cells are employed in the bioprocessing of over fifty percent of approved therapeutic proteins, attributed to their substantial growth characteristics conducive to the large-scale production of biopharmaceuticals1. Their capability for human-like post-translational modifications represents an additional advantage, which may be enhanced through gene editing techniques, as we explored in a subsequent discussion.
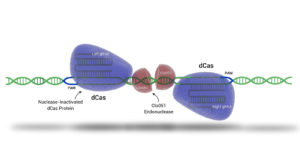
Advanced gene editing technologies, such as Demeetra's Cas-CLOVER, achieve targeted gene modification with high efficiency. This system employs two distinct guide RNAs (gRNAs) forming a pair that associate with deactivated Cas9 (dCas9) to facilitate DNA binding without cleavage. Unlike the Cas9 enzyme, DNA cutting is executed by the dimeric nuclease Clo051, thereby enhancing the precision of the edit. CRISPR-based gene editing methodologies represent a significant advancement over older gene editing tools, such as Zinc Finger Nucleases (ZFNs), which necessitated the construction of new proteins for each specific target.
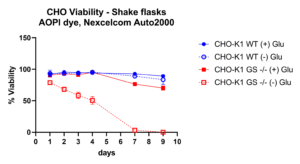
Multiplexing by co-transfection of two gRNA pairs with Cas-CLOVER resulted in a 196 base-pair deletion, eliminating nearly an entire exon of the GS gene in suspension CHO cells. Cells possessing null alleles of the GS gene (GS-/-) are viable only under conditions where an external source of glutamine is provided (denoted as ‘+ Glu’), or if they have undergone genetic modification that facilitates the stable re-expression of a functional GS gene. Here we demonstrate the large targeted deletions created by Cas-CLOVER result in a functional phenotype of GS-/- cells which die without a glutamine supply (- Glu) after seven days, while the unedited 'wild-type' cells (WT) and those with glutamine supplementation maintain viability.
Furthermore validating the functional knockout, GS CHO cell viability was recovered when the GS gene was stably transformed. Cultures in the figure were kept under +/- Glutamine conditions for 19 days.
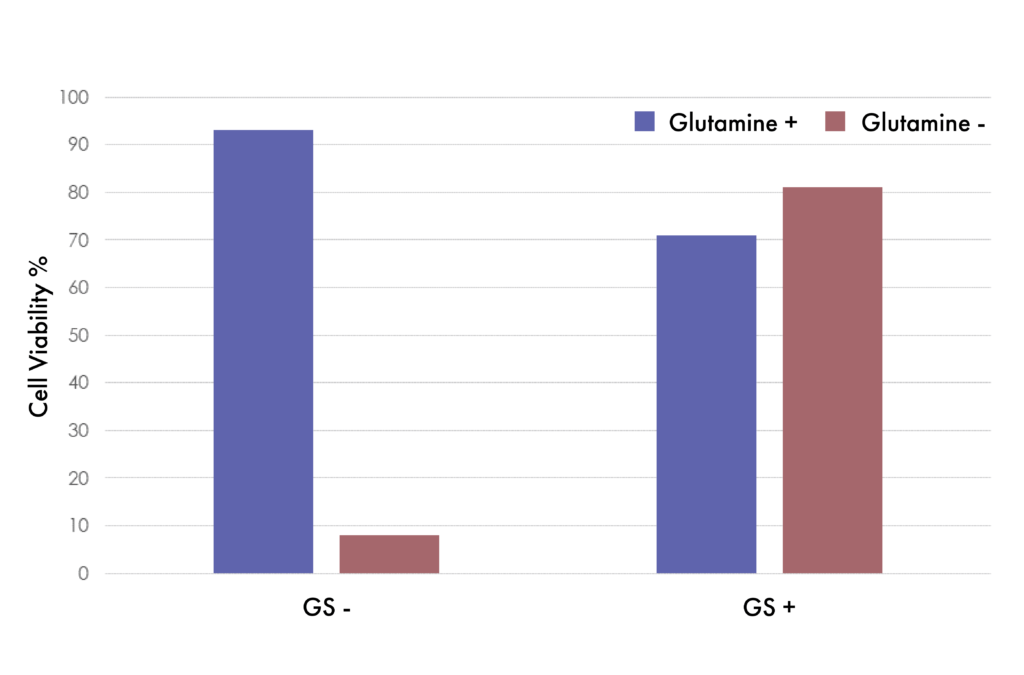
GS -/- Cells Transformed With GS Recover Viability
Suspension CHO GS -/- cells transformed with or without GS constructs. Cells not transformed with GS gene (left, GS-) lose viability without Glutamine supplementation in the media. Cells stably transformed with the GS gene (right, GS+) recover viability even without Glutamine supplementation.
In conclusion, the advancements in gene editing, have significantly bolstered the efficiency and precision of cell line development processes. The Cas-CLOVER technology, with its unique dual gRNA and Clo051 strategy, enables the creation of GS knockouts or any other gene of interest. This leap in technology not only overcomes the limitations associated with older gene-editing tools like ZFNs but also differentiates from the foundational CRISPR/Cas9 intellectual property (IP), paving the wave for clearer freedom to operate.
We encourage researchers to explore the potential of Cas-CLOVER and GS knockout CHO cells to enhance their cell line development programs. With Demeetra's exceptional technical support and expertise in gene editing, you are not just accessing tools and IP licenses but a partnership which includes proprietary protocols, know-how and one-on-one technical support from our peer-reviewed published staff. Please reach out to Demeetra to learn how our gene editing solutions can be tailored to meet your specific research and development needs.
References
- Fan (2013) Pharmaceutical Bioprocessing