Clean Gene Editing And Licensing With Cas-CLOVER
Our Cas-CLOVER gene editing technology helps eliminate off-target mutations and is available through a simple license.
Simple
Easily designed and deployed reagents

Efficient
Fast, dCas-guided sequence recognition

Specific
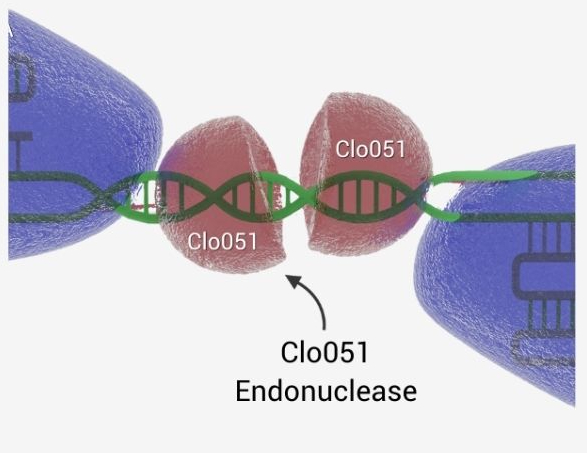
2 gRNAs match target and subunit dimerizes before cleavage
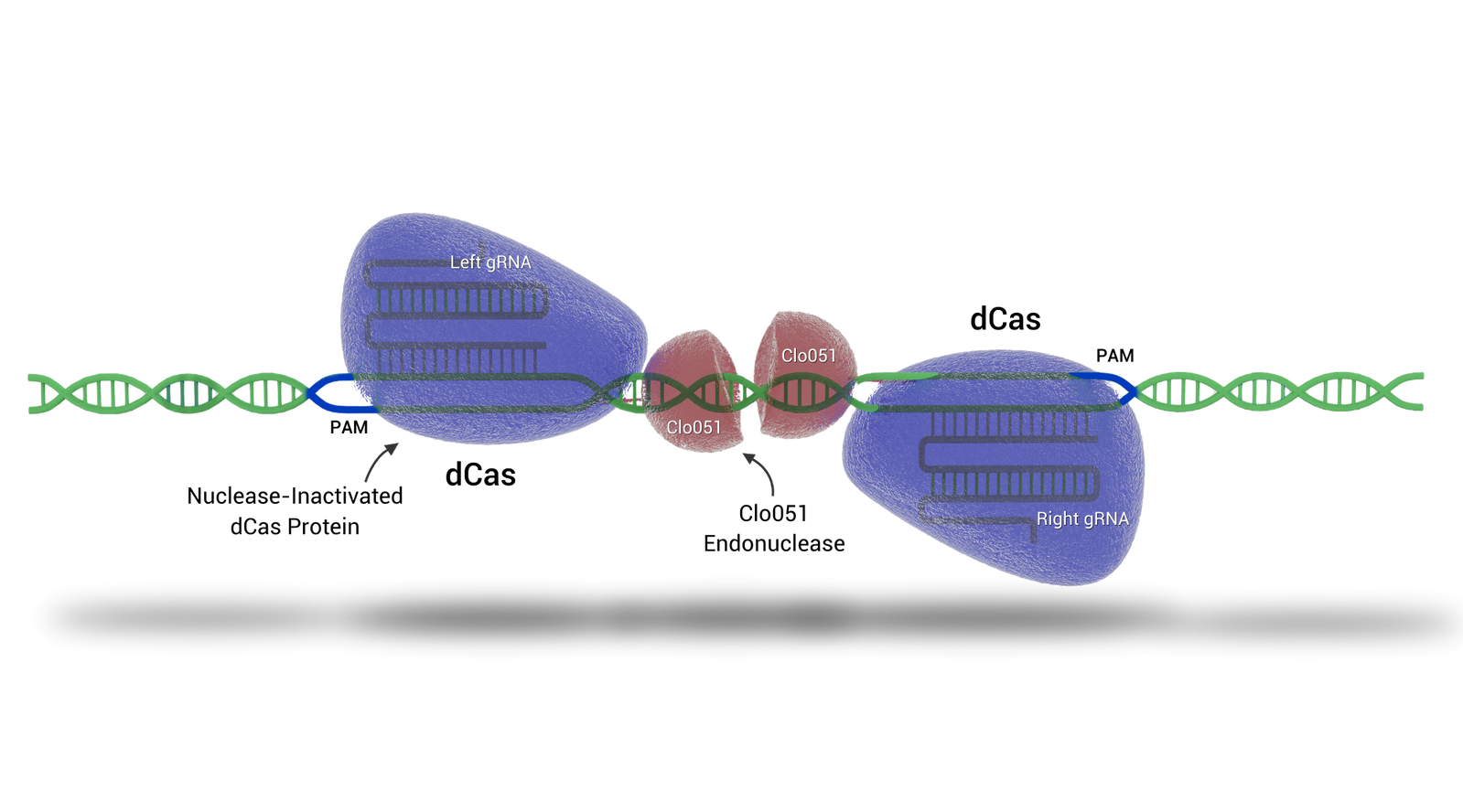
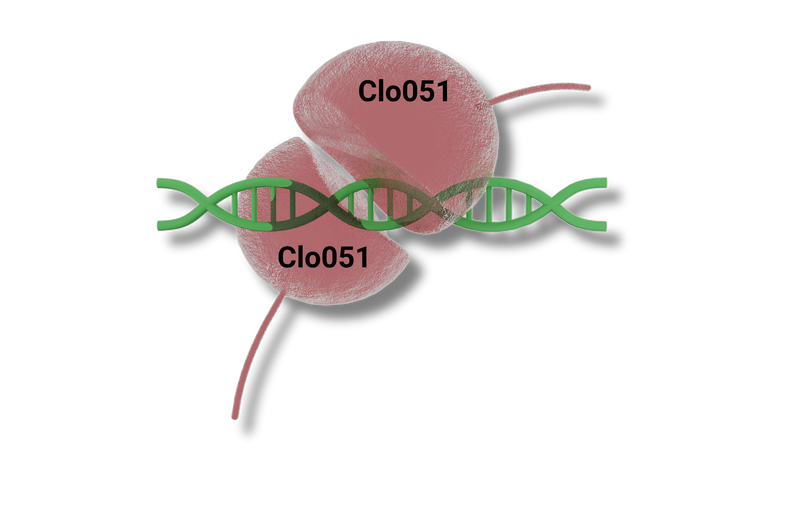
Dimerization Of Cas-CLOVER Means More Precision And High Efficiency
Cas-CLOVER utilizes the dimerization of the Clo051 nuclease to cleave DNA. The dCas fused to Clo051 is mutated so it is unable to cut DNA, serving only as a guide to the target sequence. Since Cas-CLOVER must dimerize to cut, our system requires two guide RNAs (gRNAs) to target the gene of interest, making our system highly targeted and functional only when Cas-CLOVER dimerizes at the correct target site.
Proven Efficiency in Multiple Systems
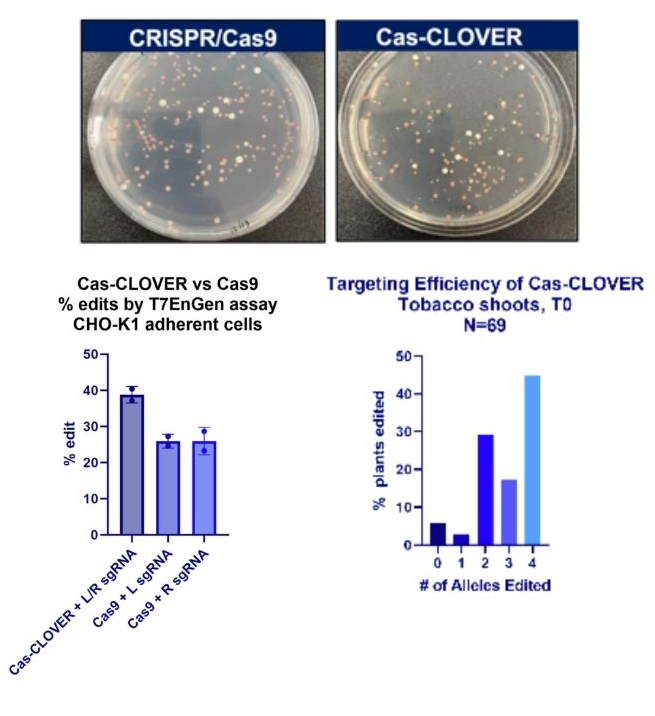
Cas-CLOVER can render full knockouts or edits in one transformation. We have demonstrated this with multiple systems in-house.
In our hands, we’ve transformed CHO cells (left graph) and S. cerevisiae (plate images, red colonies are edited) with either Cas-CLOVER or Cas9 and saw comparable editing efficiencies in both systems.
Using tobacco, a tetraploid (right graph), we’ve demonstrated Cas-CLOVER’s targeting efficiency by hitting all 4 alleles in almost 50% of regenerated shoots.
Exceptional Precision
Our Cas-CLOVER system requires two gRNAs for gene targeting, resulting in minimal off-targets. In a recent publication from our sister company, Poseida Therapeutics, potential off-target sites were identified and Cas-CLOVER demonstrated a maximum potential off-target indel rate of under 1% (Madison et al 2022).

Larger Deletions
Due to Clo051’s nuclease activity, Cas-CLOVER results in large deletions, ranging from 8 to 50 base pairs (bp). Large deletions result in more disruption of gene expression and function and enable rapid screening via traditional DNA agarose gels.
Validated In Mammalian Cells, Yeast, and Plants
We specialize in multiple industries, allowing us to apply Cas-CLOVER and piggyBac technologies into broad real world applications.

Pharmaceutical Bioprocessing
Bioprocessing and cell line engineering to produce human or non-human therapeutics
Synthetic Biotechnology
Bioprocessing and strain improvement to produce therapeutics, industrial enzymes, compounds or biofuels
Agriculture Biotechnology
Enable plant modifications that may not require GMO labels and for the production of novel therapeutics
Optimizing Your Gene Editing Research?
Contact us to learn more about our innovative gene editing technology.