The Cas-CLOVER dimeric gene editing system has demonstrated easier screening, increased rates of knockout mutations, and versatile target design capabilities. In a collaborative webinar hosted by Demeetra and Elanco Animal Health, the parties shared data pertaining to the application of Cas-CLOVER in the development of cell lines for the bioprocessing of biological therapeutics. The discussion highlighted principal advantages of the Cas-CLOVER system: its ability to streamline the screening for precise genomic editing, ensuring the achievement of complete gene knockouts, and its flexibility in gene editing target design.
Let’s take a look at the background of Cas-CLOVER, its benefits in cell line development and engineering, and how this technology is currently being utilized at Elanco.
What Is Cas-CLOVER?
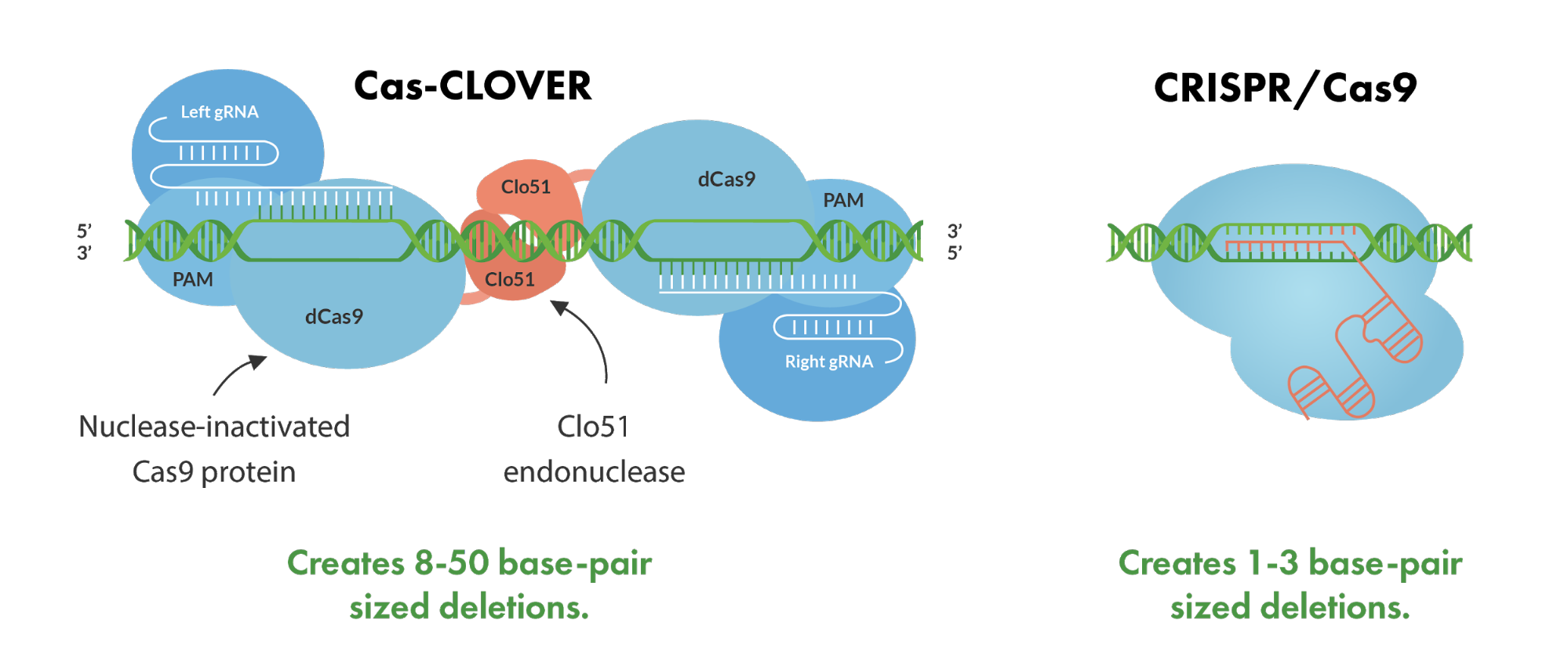
Cas-CLOVER represents a novel and effective gene-editing technology, built upon deactivated Cas (dCas) proteins fused with the dimeric Clo51 nuclease domain. Unlike the CRISPR/Cas9 system, which operates with a single guide RNA (gRNA) to direct the Cas9 enzyme for DNA cleavage, the Cas-CLOVER method employs a duo of guide RNAs. This pairing is essential for the recruitment of the editing components as monomers to the target locus. Upon successful localization at the DNA site, these monomers undergo dimerization, triggering the Clo51 nuclease domain to become active. The result is a highly specific double-strand break, featuring a 4-base pair overhang that is conducive to thorough gene knockouts and enhances the success of gene insertions or knock-ins.
The dual gRNA strategy significantly enhances the precision of the Cas-CLOVER system, markedly diminishing the prevalence of unintended or off-target effects that are a common concern with CRISPR/Cas9. The intrinsic design of Cas-CLOVER not only bolsters its editing accuracy but also carves out a distinct intellectual property niche within the increasingly crowded genome editing landscape. Hence, Cas-CLOVER stands out as a simple yet potent tool for targeted genome engineering, capable of efficiently modifying a vast array of bioprocessing cell lines with unprecedented fidelity.
Advantages Of Cas-CLOVER System
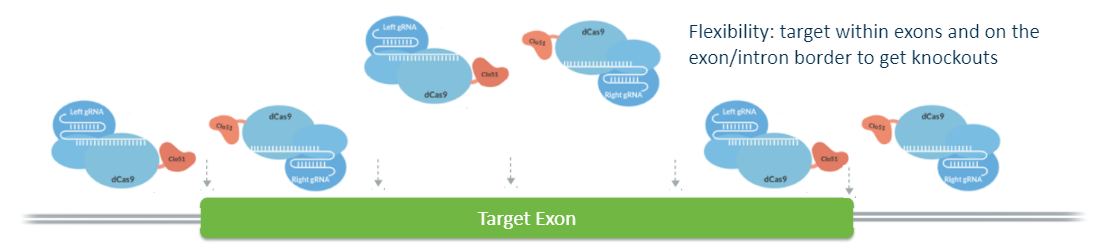
A significant benefit is facilitated by the flexible spacer lengths available in the dual gRNA approach, expanding the range of accessible targets. Scientists determined that optimal activation of the Cas-CLOVER system, occurs when two guide RNAs (gRNAs) are spaced 11 to 31 base pairs apart. The utility of this spacer range extends to various cell types, including those for bioprocessing, and to diverse organisms such as plants and yeast. The flexibility in spacer length paired with the necessity for dual PAM sites enhances Cas-CLOVER's applicability for precise gene targeting, leveraging the simplicity of CRISPR technology.

Experimental validation of the gRNA spacer in HEK293 cells utilized a construct for assessing activity through GFP expression upon DNA cleavage by Cas-CLOVER.
When looking at Cas-CLOVER’s deletion performance compared to the current standard technologies, Cas-CLOVER shows that it has clear advantages. The Cas-CLOVER system creates larger deletions in the 8-50 bp range in comparison to CRISPR/Cas9 which typically generates 1-3bp deletions.
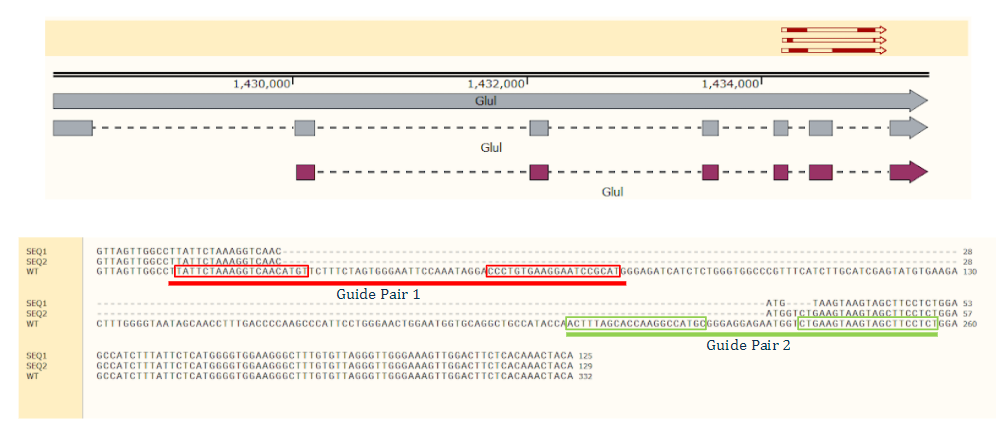
Top: The sequence alignment of indels. Bottom: TOPO clone indel sequence alignment from Cas-CLOVER mediated targeting of Single Celled Clones reveal deletions ranging from 13 to 25 bp deletions. All deletions create a frameshift mutation that will eliminate glutamine synthetase protein expression.
Why Does This Matter To You?
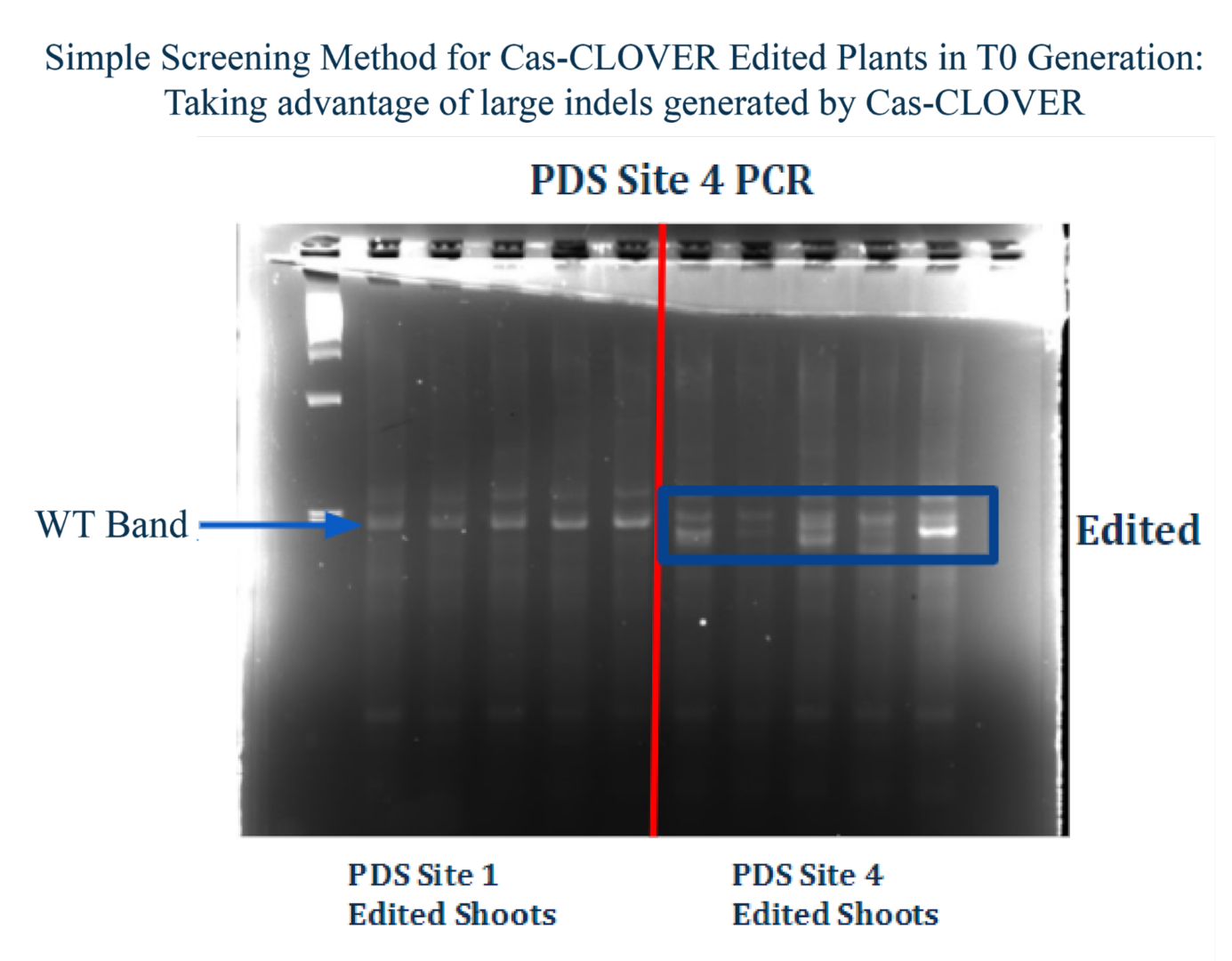
Larger deletions mean easier screening – Unlike the smaller deletions created by Cas9, Cas-CLOVER edited cells and plants can be positively identified by running quick and easy gels. Gel based indel identification can be run in-house and within a matter of hours, reducing costs, streamlining workflows and avoids sending samples out of the lab for sequencing such as next-generation sequencing (NGS).
The image above demonstrates a simplified screening technique for targeted gene knockouts utilizing the Cas-CLOVER system. In this study, leaf samples were taken PDS Site 4 edited plants, and genomic DNA was then extracted via NaOH alkaline lysis. Genome editing was confirmed by PCR, followed by gel electrophoresis. Importantly, the individual conducting the experiment was blinded of the sample identities, specifically which lanes contained edited specimens or wild-type (WT) controls. This study contrasts with the outcomes typically observed with the CRISPR/Cas9 system, where smaller deletions may not consistently achieve complete gene expression knockout. In our recent presentation, we demonstrate that Cas-CLOVER can delete entire exons in CHO cells. Let’s review the details below.
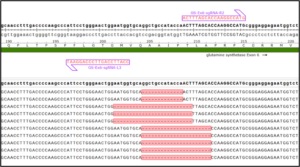
Knock-Out And Full Deletion Of The Entire Exon In GS In Suspension CHO Cells
GS knock-out was performed in CHO cells suspension, along with a full deletion of the entire Exon in GS. Guides targeting the exon/intron boundaries of an entire Exon in GS resulted in full exon deletion, determined by NGS amplicon deep sequencing.
Cas-CLOVER is naturally dimeric and requires two guide RNAs, meaning it may require a little more design on the front end, but the downstream benefits are well worth it. The system ultimately creates larger indels, so one of the guides can be designed outside of the target exon. You can also design the deletion to encompass the adjacent exon resulting in a successful knockout mutation.
Interested In Evaluating Cas-CLOVER?
We cordially invite the scientific community to explore the potential of Cas-CLOVER technology. Through Demeetra's unparalleled expertise in gene editing and our extensive technical support framework, researchers are afforded access to not only state-of-the-art tools and intellectual property licensing options but also a synergistic partnership. This partnership encompasses proprietary protocols, specialized expertise, and dedicated assistance from our team of experts distinguished by their contributions to peer-reviewed research.