Browse Our Powerful Gene Editing Toolbox Of Precision Cas-CLOVER™ And Flexible piggyBac® Gene Insertion Technologies
Cas-CLOVER and piggyBac reagents are readily available for mammalian, plant, and yeast. Reach out to us to learn more about our evaluation and commercial license options with full freedom to operate.
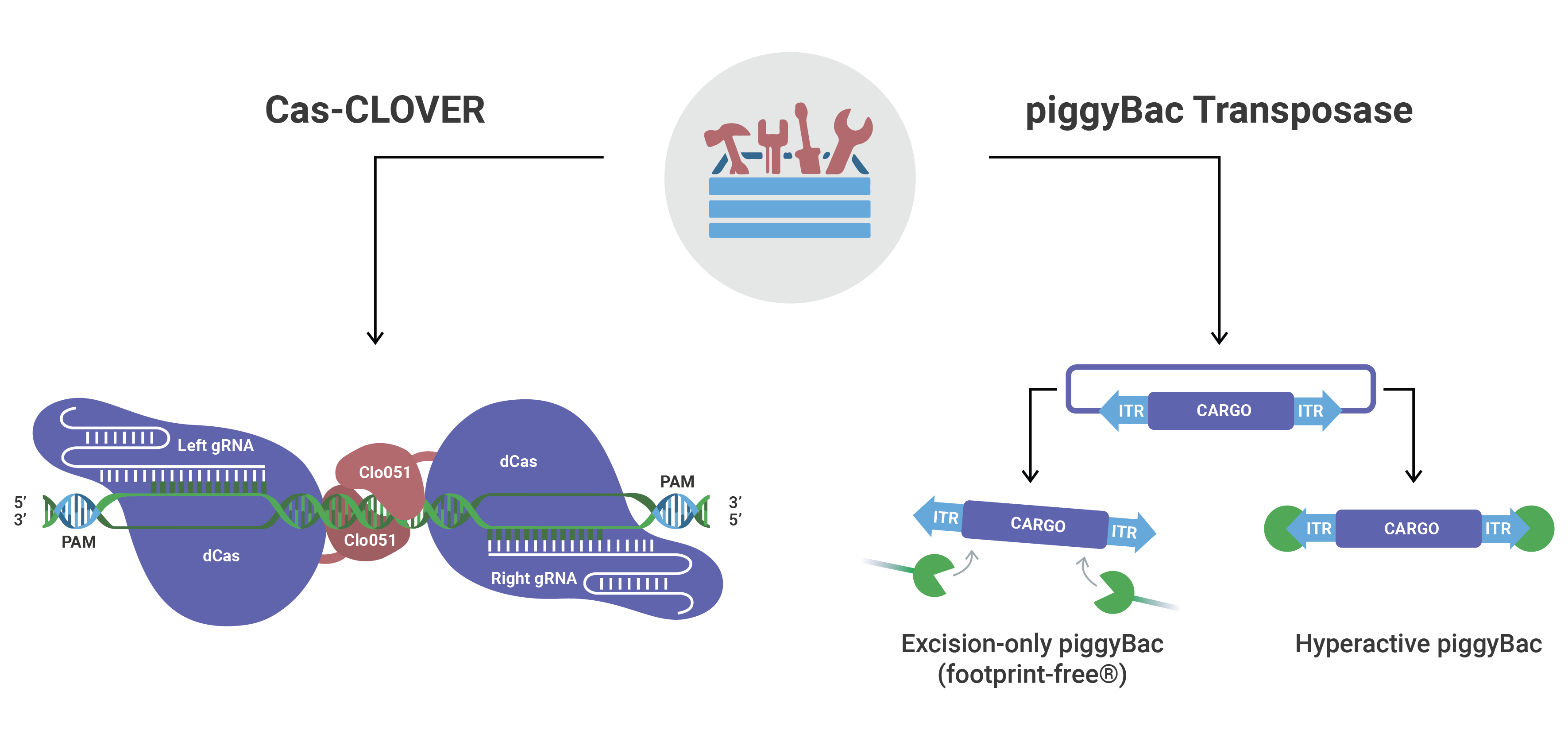
Mammalian Gene Editing
| SKU# | Technology | Description | Resource |
|---|---|---|---|
| rDMT5-011 | Cas-CLOVER | Cas-CLOVER mRNA: Codon optimized research grade mRNA for mammalian systems. Order gRNAs from your favorite provider and transform with the mRNA for high efficiency and clean editing. Components: 1 vial containing 25 µg (1 mg/mL) of mRNA | Design Guide |
| rDMT5-027 | piggyBac | Super piggyBac transposase mRNA: Codon optimized research grade mRNA for mammalian systems. Transform cells with piggyBac mRNA and piggyBac transposon (pDMT5-028) for stable integration. Components: 1 vial containing 25 µg (1 mg/mL) of mRNA | User Guide |
| rDMT5-020 | piggyBac | Excision-only piggyBac transposase mRNA: Use Excision-only piggyBac transposase to remove previously integrated piggyBac transposons for phenotype reversions and Footprint-Free gene editing. Components: 1 vial containing 25 µg (1 mg/mL) of mRNA | User Guide |
| pDMT5-029 | piggyBac | Super piggyBac transposase plasmid: Codon optimized for mammalian systems. Transform cells with piggyBac plasmid and piggyBac transposon (pDMT5-028) for stable integration. Please note this plasmid is not expandable. Components: 1 vial containing 25 µg of plasmid DNA at 1 mg/mL | Map User Guide |
| pDMT5-030 | piggyBac | Excision-only piggyBac transposase DNA: Use Excision-only piggyBac transposase to remove previously integrated piggyBac transposons for phenotype reversions and Footprint-Free gene editing. Please note this plasmid is not expandable. Components: 1 vial containing 25 µg of plasmid DNA at 1 mg/mL | Map User Guide |
| pDMT5-028 | piggyBac | piggyBac transposon: Using the flexible MCS, clone in your promoter, gene and other components for optimal expression in your system. This plasmid is expandable. Components: 1 vial containing 10 µg of plasmid DNA | Map |
Plant Gene Editing
| Plasmid Sku# | Codon Optimization | Cas-CLOVER Promoter | gRNA Promoter | Host Selection | Bacterial Selection | Map link |
|---|---|---|---|---|---|---|
| pDMT1-1 | A. thaliana | CaMV35S | OsU3 | Hygromycin | Kanamycin | Map |
| pDMT3-4 | A. thaliana | CaMV35S | AtU6-26 | Hygromycin | Kanamycin | Map |
| pDMT1-123 | A. thaliana | CaMV35S | OsU6 | Hygromycin | Kanamycin | Map |
| pDMT1-112 | A. thaliana | CaMV35S | OsU3 | Kanamycin | Kanamycin | Map |
| pDMT1-114 | A. thaliana | CaMV35S | AtU6-26 | Kanamycin | Kanamycin | Map |
| pDMT1-116 | A. thaliana | CaMV35S | OsU3 | Glufosinate | Kanamycin | Map |
| pDMT1-118 | A. thaliana | CaMV35S | AtU6-26 | Glufosinate | Kanamycin | Map |
Yeast Cas-CLOVER Reagents
| Plasmid Sku# | Codon Optimization | Cas-CLOVER Promoter | gRNA Promoter | Host Selection | Host Replication | Bacterial Selection | Map link |
|---|---|---|---|---|---|---|---|
| pDMT1-124 | S. cerevisiae | ScRNR2 | No Guide Expression Cassette | LEU2 | CEN/ARS | Ampicillin | Map |
| pDMT1-125 | S. cerevisiae | ScRNR2 | ScSNR52 | LEU2 | CEN/ARS | Ampicillin | Map |
| pDMT1-126 | S. cerevisiae | ScRNR2 | ScSNR52 | URA3 | CEN/ARS | Ampicillin | Map |
| pDMT1-127 | S. cerevisiae | ScRNR2 | ScSNR52 | KanR | CEN/ARS | Ampicillin | Map |
| pDMT1-128 | S. cerevisiae | ScRNR2 | ScSNR52 | HygR | CEN/ARS | Ampicillin | Map |
Yeast piggyBac Reagents
| Plasmid SKU# | Codon Optimization | Description | Map link |
|---|---|---|---|
| pDMT3-23 | S. cerevisiae | Taking advantage of the unique massive cargo capacity, this ~25kb piggyBac transposon vector contains all genetic components required for engineering the yeast mevalonate pathway (MVA) in a single-step for the production of a high value secondary metabolites. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-1 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-1 has a native URA3 promoter driving a URA3 auxotrophic marker. This plasmid is expandable Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-2 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-2 has a defective URA3 promoter driving a URA3 auxotrophic marker allowing for more potential integrations of the transposon cassette. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-3 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-3 has a native LEU2 promoter driving a LEU2 auxotrophic marker. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-4 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-4 has a defective LEU2 promoter driving a LEU2 auxotrophic marker allowing for more potential integrations of the transposon cassette. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-5 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-5 has a native HIS3 promoter driving a HIS3 auxotrophic marker. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-6 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-6 has a defective HIS3 promoter driving a HIS3 auxotrophic marker allowing for more potential integrations of the transposon cassette. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-7 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-7 has a native TRP1 promoter driving a TRP1 auxotrophic marker. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
| pDMT4-8 | S. cerevisiae | Customizable piggyBac transposon vector containing a large multiple cloning site (MCS) flanked by piggyBac ITRs for integration and core insulators which protect against gene silencing. pDMT4-8 has a defective TRP1 promoter driving a TRP1 auxotrophic marker allowing for more potential integrations of the transposon cassette. This plasmid is expandable. Components: 1 vial containing 10µg of plasmid DNA | Map |
