The food industry is undergoing a significant transformation with the rise of animal-free meat alternatives, including cell-based meats and plant-based proteins. According to a report by the Boston Consulting Group, this burgeoning market is expected to soar to a valuation of $290 billion by 2035, accounting for 11% of the total protein intake globally. Initially, the industry focused on plant-based meats that replicate the taste of meat using heme proteins, alongside a blend of microbial and plant proteins, to achieve a texture and appearance close to that of actual meat. This approach has garnered considerable success, paving the way for the next innovative leap: creating meat cells and proteins that not only resemble but also match the nutritional value of traditional meat, all without involving animals.
Cell Line Engineering may be the Future of Cultured Meat
This next wave of progress in alternative foods is upon us with the advent of lab-grown meats and seafoods, derived from cultured cell lines. This innovative approach involves cultivating cells, which can be initially sourced through a simple biopsy, in a lab setting to produce genuine animal-based foods. Companies globally are at the forefront of this exciting trend, working to create products that go beyond merely mimicking the appearance of traditional meats and seafoods.
One of the challenges in this process is to generate cell lines that are renewable or stem cell-like to remove the need for more samples and to maintain consistency. These cells can be manipulated to improve their growth in a laboratory or bioreactor; differentiate into specific tissues such as muscle; and enhance production of certain protein(s) for optimal nutritional composition, appearance and texture. By optimizing culture conditions and differentiation protocols using engineered cells, researchers have unlocked the potential to enhance efficiency and increase yields, thereby reducing overall production costs. A key innovation detailed in a recent study is the development of engineered autocrine signaling systems, particularly the expression of Fibroblast Growth Factor 2 (FGF2), which greatly reduces the need for costly media supplements traditionally required for cell growth and differentiation. Engineered cells were designed to express FGF2, thereby creating self-sustaining cell proliferation and differentiation with minimal reliance on external growth factors1.
Using PiggyBac for Faster Results
Engineering cell lines is notoriously slow compared to microbial cells. However, a tool like Demeetra's piggyBac transposase system allows quick improvement of mammalian as well as seafood cell lines in an extremely short time compared to traditional methods (Figure 1).
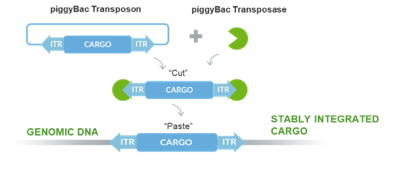
Figure 1: PiggyBac stable transposition mechanism. DNA “Cargo” including immortalization genes, differentiation factors or genes expressing proteins of interest, are cloned into transposon vector. Co-transfection of the transposon(s) and transposase mRNA into cells cuts out cargo and pastes into random TTAA sites which can be screened for the desired insert or trait.
PiggyBac can generate stably expressed proteins at a very high efficiency and allows researchers in the alternative protein space to quickly screen for cells with desired characteristics. Insertion preference into highly-expressed genomic loci is a unique characteristic of piggyBac that has been utilized with great success in the biopharmaceutical industry for cell immortalization, reprogramming, and differentiation, as well as the production of proteins. When piggyBac is used for antibody bioproduction, it yields up to more than 11-fold increases in comparison with standard plasmid transfection2.
Addressing Roadblocks to Meat Cell Proliferation
Meat cells have minimal lifecycles in culture, limiting large scale propagation. Maintaining proliferating cells without affecting their ability to differentiate into lineages such as muscle, is a critical step in cultured meat production. One way to accomplish this is to introduce genes that result in cells with unlimited proliferation capacity. PiggyBac is particularly useful for this purpose because it integrates stably at highly expressed sites, thereby enabling large scale propagation without loss as with transient expression.
Another consideration is the DNA cargo capacity for integration of the desired genes. When one group attempted retrovirus-mediated immortalization of mouse embryonic fibroblasts (MEFs), little efficiency was observed due to low viral titers of the large cargo required. When the same group used piggyBac’s massive cargo capacity, they efficiently immortalized mouse MEFs while maintaining multipotency.
As demonstrated in Figure 3 below, primary MEFs (top panel a) grew slowly through five passages (P5) but piggyBac immortalized MEFs (piMEFs, lower panel b) grew rapidly and through at least forty passages (fifteen shown, P15). Further, a cell staining assay shows that piMEFs reach confluence on day 3 versus day 7 for primary MEFs and had significantly higher cell numbers measured as optical absorbance at all timepoints (Figure 3b)3.
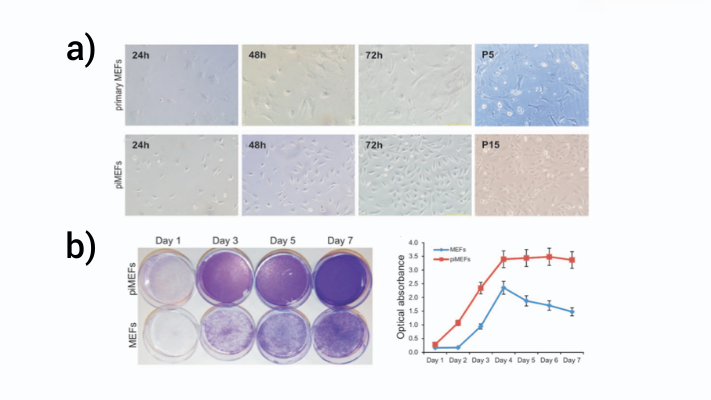
Figure 3: PiggyBac stable expression of SV40T immortalizes MEFs.
Optionally, following the transposon mediated insertion, the cassette can be removed in a scarless manner with excision-only piggyBac transposase (PBx) as shown below in Figure 4. We call this process Footprint-Free gene editing. This useful feature of piggyBac can be applied to removing reprogramming or immortalization factors to drive differentiation following proliferation, phenotype reversion and selection marker recycling.
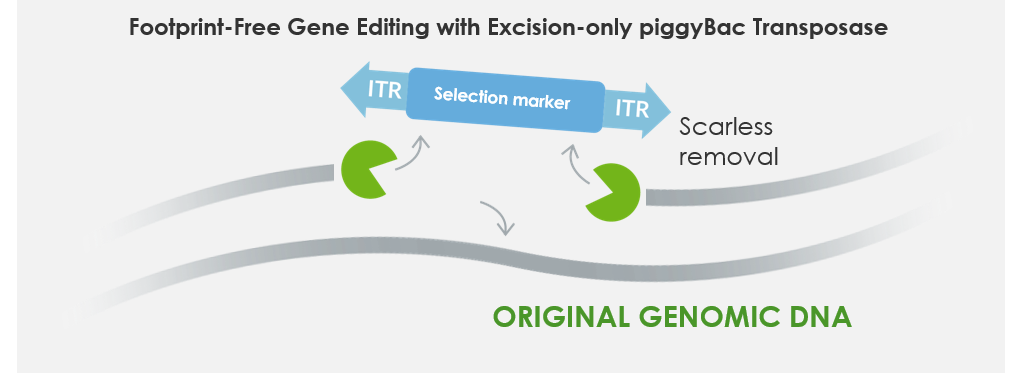
Figure 4: Footprint-Free gene editing. Excision-only piggyBac transposase mRNA removes the stably inserted transposon.
Why Use Demeetra’s Gene Editing Platform?
Understandably, many companies do not want to invest too heavily to establish new basic protocols. So, the final question one might ask is, has it been used in my system or species? This is where piggyBac excels as it is active in nearly all species and cells tested to date. See piggyBac’s abundant applications and uses below.
| Species | Outcome | Reference link |
|---|---|---|
| Chicken | optimization in chicken primordial germ cells (PGCs) | Xian et al. 2021 |
| Cow | piggyBac outperforms other transposons in transgenic cattle production | Yum et al. 2016 |
| Pig | triple transgenic pigs for enhanced feed utilization | Wang at al. 2020 |
| Fish | piggyBac activity confirmed in three species of fish | Hu et al. 2013 Shen et al. 2018 |
| Mollusks | transgenesis in Pacific oysters, including stable insertion of growth hormone | Chen et al. 2018 |
| Goat | piggyBac transgenesis enhances cashmere yield in goats | Shi et al. 2017 |
Further Optimizations and the Role of Targeted Nucleases
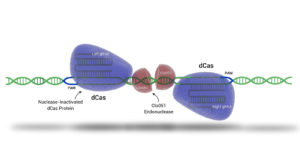
In addition to piggyBac, Demeetra's Cas-CLOVER can be used to generate a more precise deletion/insertion once genomic areas of interest are identified with piggyBac (Figure below). Cas-CLOVER and piggyBac, can alternatively be used in conjunction to generate footprint free mutants. These targeted approaches enable researchers to fine-tune cellular functions, improve growth kinetics, and optimize differentiation pathways, ultimately contributing to a more cost-effective and sustainable production process.
Learn More About Demeetra
Our flagship gene editing systems, Cas-CLOVER and piggyBac, offer enormous advantages and flexibility. When working with our technologies, research teams benefit from faster and more precise development of products. Contact us to learn more about our independent gene editing technology IP, optimized reagents and internal expertise to help guide smooth adoption.
References
- Stout et al. (2024) Engineered autocrine signaling eliminates muscle cell FGF2 requirements for cultured meat production. Cell Reports Sustainability.
- Rajendra et al. (2016) Generation of Stable Chinese Hamster Ovary Pools Yielding Antibody Titers of up to 7.6 g/L Using the piggyBac Transposon System. Biotechnol. Prog., Vol. 32, No. 5
- Wang et al. (2016) The piggyBac Transposon-Mediated Expression of SV40 T Antigen Efficiently Immortalizes Mouse Embryonic Fibroblasts (MEFs). Plos One.