Using Unbiased Analysis Cas-CLOVER’s Maximum Potential Off-target Is Under 1% Where CRISPR/Cas9 Is 5-13%
Detailed investigations in gene-editing technologies have highlighted the critical importance of target specificity. Within this context, Cas-CLOVER emerges as a exceptional alternative to the CRISPR/Cas9 platform, offering a more accurate solution for gene editing. Cas-CLOVER distinguishes itself from the CRISPR/Cas9 system through its remarkable specificity and minimal off-target effects. Comparative analyses have demonstrated that Cas-CLOVER can achieve indel frequencies comparable to those of CRISPR/Cas9, yet with significantly reduced off-target mutagenesis.
The system’s enhanced specificity is evidenced by exhaustive off-target analyses, which employ advanced algorithms to identify potential off-target sites based on sequence similarity. Subsequent deep sequencing of these sites, utilizing next-generation sequencing (NGS) techniques, has consistently revealed an absence of unintended mutations, affirming Cas-CLOVER’s precision and reliability. Below are a few figures representing Cas-CLOVER’s effectiveness at preventing off-target activities1.

Top: possible combination of gRNAs. Bottom: examples of prediction results for possible off-target locations fpr gRNA combination L+R and R+R.
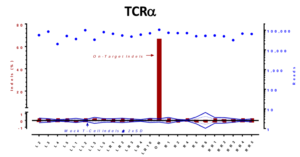
On-target versus potential off-target mutagenesis in T-cells. Genomic DNA was collected and the top predicted off-target sites were selected for target and sequencing by NGS. No off-target mutations above background were observed by NGS sequencing, with most of the sites at 30,000 to 100,000 X coverage.
Cas-CLOVER has also been used to create human cell therapy product candidates by our sister company, Poseida Therapeutics. In addition to the NGS work, extensive off-target analysis has demonstrated essentially no off-target cutting above background frequency.
In the image below, Cas-CLOVER edited T-cells from seven donors were analyzed by iGuide-seq followed by Ampli-seq. After filtering out background, only 19 potential off-target sites had detectable editing. Target sites 1 & 2 below have no homology to the target site gRNAs and may be false positives. The rest of the sites show under 0.1% indel rate in all donors, this nearly undetectable level is present while the on-target cutting rates were 92-99%2.
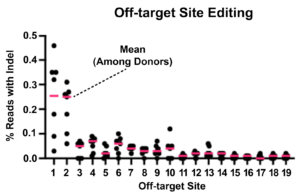
Off-target-site-editing averages were between 0.012 – 0.089%, when false positive targets 1 & 2 are eliminated.
In contrast to Cas-CLOVER, the monomeric CRISPR/Cas9 nuclease system is directed to target sites with a single gRNA. Cas9 retains the capability to bind to and cleave DNA sequences even in the presence of several base-pair mismatches within the guide RNA (gRNA), thereby elevating the potential for considerable off-target mutagenesis. In Chinese Hamster Ovary (CHO) cells, the employment of single gRNAs with either one or two base-pair mismatches was observed to lead to an off-target insertions and deletions (indels) frequency ranging from 1% to 5%. Notably, one specific Cas9 gRNA was associated with an off-target mutation frequency of 10.7%. Furthermore, analysis of off-target sites for another gRNA revealed the existence of off-target mutations in three out of four sites examined.
Learn More About Cas-CLOVER And Demeetra
In summary, the Cas-CLOVER technology represents a significant advancement in gene editing, prioritizing specificity and minimizing off-target effects, thereby setting a new standard for precision in genetic modifications. Partnering with Demeetra cuts through the conventional bounds of acquiring tools and intellectual property licenses. We seek a collaborative relationship enriched with exclusive protocols, expertise, and personalized technical support provided by our peer-reviewed staff. We invite you to engage with Demeetra and explore how our gene editing solutions can be adopted to fulfill your unique research and development objectives.
References
1: Li et al (2018) Cas-CLOVER™: A High-Fidelity Genome Editing System for Safe and Efficient Modification of Cells for Immunotherapy. Precision CRISPR Congress Poster.
2: Madison et al. (2023) Cas-CLOVER is a novel high-fidelity nuclease for safe and robust generation of TSCM-enriched allogeneic CAR-T cells. Molecular Therapy Nucleic Acids.
3: Lee et al (2015) Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway. Scientific Reports.
