Surgical Synthetic Biotechnology: Editing Yeast With Cas-CLOVER & piggyBac transposase
Our genome editing toolbox integrates the precise cutting capability of Cas-CLOVER with the piggyBac transposase's capacity to stably integrate large genetic cargo and facilitate seamless removal of transgenes, positioning it at the forefront of synthetic biotechnology. This document will examine the mechanisms by which these systems operate, highlight their distinct features, and compare them with similar technologies in the field.
The Cas-CLOVER System & Its Benefits
Cas-CLOVER is a gene editing system that shares many benefits with CRISPR/Cas9 but has distinct advantages due to its unique components. Unlike CRISPR/Cas9, which uses a single guide RNA (gRNA) and the Cas9 enzyme to cut DNA, Cas-CLOVER employs two separate gRNAs and a deactivated Cas9 (dCas9) for DNA binding without cutting. Instead of Cas9, the cutting action is performed by the dimeric nuclease Clo051, enhancing the system's precision and significantly reducing unwanted and off-target mutations. Additionally, Cas-CLOVER creates larger 4-base pair overhang deletions, which can lead to more complete gene knockouts and improved efficiency and size for gene insertions (knock-ins).
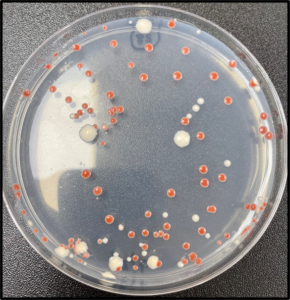
In the yeast, Saccharomyces cerevisiae (S. cerevisiae), deleting the ade2 gene exhibits a red colony color phenotype through intermediate metabolite accumulation of the adenine biosynthesis pathway. Using Cas-CLOVER, we successfully integrated a homology vector at the ade2 locus, thereby achieving its complete gene deletion. This simple colony count assay was utilized as a method for testing gene editing efficiencies.
How piggyBac Transposase Works
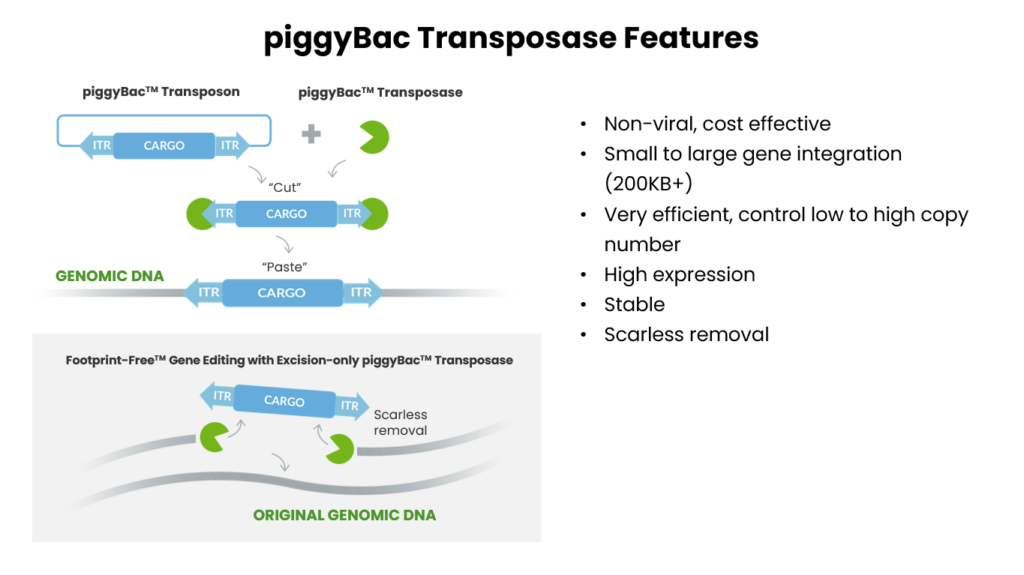
One of the most versatile and widely adopted transposon systems is the piggyBac transposase platform, which is known for providing insertional mutagenesis or stable expression and precise genome engineering through seamless excision. Specifically, piggyBac is a TTAA-specific, cut and paste transposase that’s shown to function efficiently independent of host cell factors, as well as having a high propensity for integration into highly transcribed units, making it a superior tool for insertional mutagenesis for most applications.
The transposon piggyBac is being used increasingly for genetic studies. One study describes the modified versions of piggyBac transposase that have potentially wide-ranging applications, such as reversible transgenesis and modified targeting of insertions. piggyBac is distinguished
by its ability to excise precisely, restoring the donor site to its pre-transposon state. This characteristic makes piggyBac useful for reversible transgenesis, a potentially valuable feature when selectable marker recycling or phenotype reversion is desired. [1]
Additionally, the piggyBac transposase is very versatile and has the capabilities of very large cargo sequences, with capacities of 200kb+ at high frequencies and across a vast array of organisms, including all yeast species ever attempted. In contrast to most other transposon systems, piggyBac doesn’t have any overproduction inhibition, making deployment in new hosts very simple by enabling the use of specialized promotors that are commonly used for gene overexpression. Furthermore, piggyBac is also capable of efficient simultaneous integration of multiple different DNA sequences. [2]
Combining Cas-CLOVER And piggyBac
There have been multiple studies validating the effectiveness of combining Cas-CLOVER & piggyBac technologies to create “Footprint Free” gene editing. With this combination the options are endless, from metabolic engineering and industrial manufacturing to genome editing in yeast using Cas-CLOVER & piggyBac.
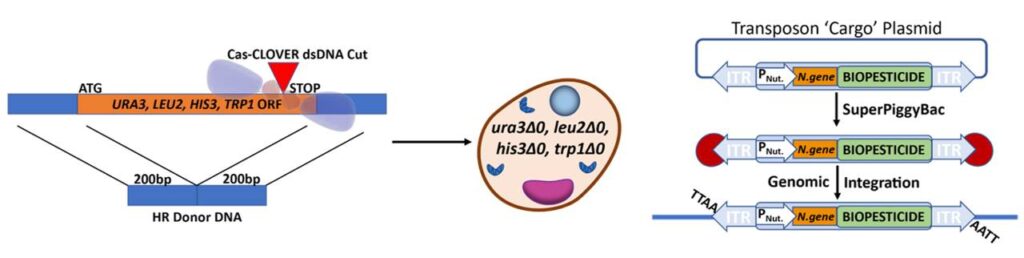
One example is the development of yeast strains for industrial-scale applications in RNAi yeast-based mosquito control biopesticides. Cas-CLOVER was used to engineer yeast strains with multiple auxotrophic gene deletions, such as ura3 and leu2 (Figure 1A). These gene edited strains can be complemented by wild-type nutritional markers, which facilitates the selection of transgenic lines containing inserted cargo, in this case biopesticide RNAi, and selectable genes. This process employs piggyBac transposase/transposon technology to enable positive selection and efficient detection of integrated cargo (Figure 1B).
A Simple, Versatile & Efficient Tool For Yeast Strain Development
One of the biggest benefits to working with piggyBac in yeast development is that it’s potentially active in all yeast strains, including difficult to engineer yeasts. One piggyBac vector can rapidly develop mutant libraries for loss (gene-trapping) and gain of function (enhancer trapping) across all species, thus creating an efficient platform. With piggyBac, genes can be easily mapped, and phenotype reversion is a completely seamless activity. The piggyBac system has the capabilities of expediting the screening process for multiple commercial yeast strains all within a single transformation step, and even without any preconceived knowledge of the strain’s genetics or target site sequences – making piggyBac a huge plus!
Research teams can potentially screen thousands of commercial strains to determine if they are amenable to metabolic engineering, and tolerant of the end product.
References
- Xianghong Li, Erin R. Burnight, Ashley L. Cooney, Nirav Malani, Troy Brady, Jeffry D. Sander, Janice Staber, Sarah J. Wheelan, J. Keith Joung, Paul B. McCray Jr., Frederic D. Bushman, Patrick L. Sinn, and Nancy L. Craig. piggyBac transposase tools for genome engineering. PNAS June 18, 2013 110 (25) E2279-E2287; https://doi.org/10.1073/pnas.1305987110
- Wagner, J., Williams, E., Alper, H. (2018). Developing a piggyBac transposon system and compatible selection markers for insertional mutagenesis and genome engineering in Yarrowia lipolytica. McKetta Department of Chemical Engineering, The University of Texas at Austin. https://doi.org/10.1002/biot.201800022
- Brizzee, C., Mysore, K., Njoroge, T. M., McConnell, S., Hamid-Adiamoh, M., Stewart, A. T. M., Kinder, J. T., Crawford, J., & Duman-Scheel, M. (2023). Targeting mosquitoes through generation of an insecticidal RNAi yeast strain using Cas-CLOVER and Super PiggyBac engineering in Saccharomyces cerevisiae. Journal of Fungi, 9, 1056. https://doi.org/10.3390/jof9111056