Using a single ~29kb vector, Demeetra’s proprietary Super piggyBac transposase stably introduced the entire 11-gene mevalonate pathway, a key metabolic component manufacturing synthetic biology (SynBio) products in s. cerevisiae. This approach can be applied to any pathway in any yeast strain without preconceived knowledge of the target site for homologous recombination.
From industrial biotechnology to agriculture and beyond, the piggyBac transposase is a versatile tool that is becoming increasingly utilized in a wide variety of applications. In particular, researchers have adopted a widespread use of piggyBac for cell line development with Chinese Hamster Ovary (CHO) cells, which are in turn used for the bioprocessing of high value proteins and therapeutics, such as monoclonal antibodies (mAbs).
Researchers at Demeetra have demonstrated that the piggyBac platform possesses the capability to integrate a substantial number of transgenes across various host organisms, boasting an almost limitless cargo capacity. The piggyBac transposase exhibits a propensity to integrate stably within highly transcribed genomic regions, thereby serving as an invaluable tool for the development of cell lines and strains that express genes of interest at high levels.
This discussion will delve deeper into how the piggyBac's substantial genetic cargo delivery facilitates efficient metabolic engineering of yeast, ultimately enabling the production of high-value metabolites and other synthetic biology products.
Mevalonate Pathway Combined With piggyBac Transposon
To demonstrate piggyBac system efficiency in SynBio and metabolic engineering, we engineered a large pathway in Saccharomyces cerevisiae (S. cerevisiae) yeast.
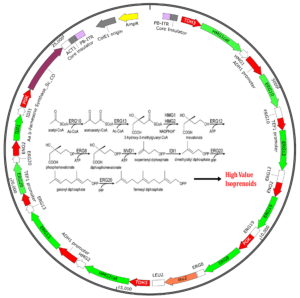
β-Farnesene is produced by a complex metabolic pathway involving 11 genes using a single transposon with piggyBac transposase.
The focus of these proof-of-concept studies was to engineer the yeast mevalonate pathway (MVA) in order to optimize the production of a high value sesquiterpene, called β-Farnesene. Our researchers rationalized that by demonstrating the delivery and expression of all genes in the yeast pathway on a single piggyBac cassette, would provide significant value to the SynBio field for producing high-value secondary metabolites.
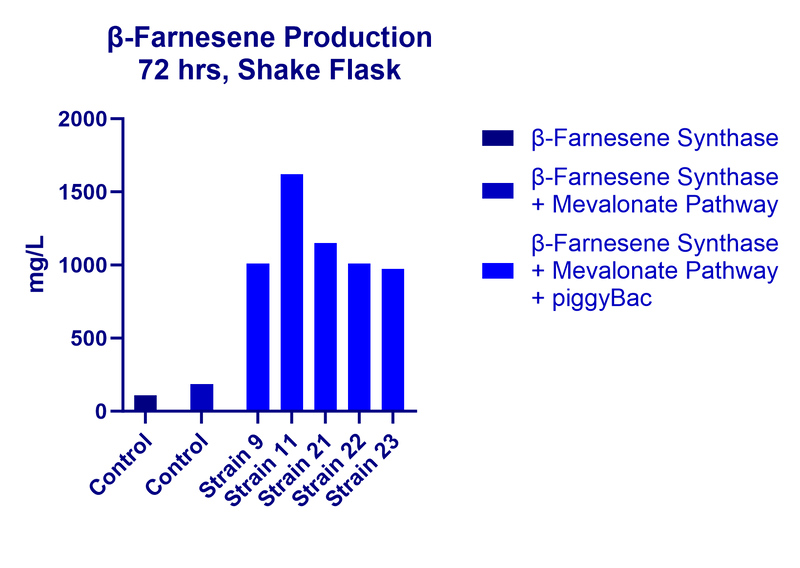
β-Farnesene Production After Single Plasmid Transformation Of S. cerevisiae Strain
In this study, our team transformed the entire S. cerevisiae MVA pathway within a single piggyBac cassette, along with a β-Farnesene synthase. In a single transformation of the piggyBac/MVA cassette, we were able to obtain multiple strains with high levels of β-Farnesene production. When these high-producing strains were carried over to a shake flask experiment, the highest titer reached in a given strain was over 1.6 g/L.
Ultimately, these proof-of-concept results show that the piggyBac system has a very robust performance in S. cerevisiae.
Pros of piggyBac System For Yeast Strain Metabolic Engineering
The study results above elucidate the approach to implementing the piggyBac system for improving the production of metabolites in transformed S. cerevisiae. These results clearly demonstrate that the PiggyBac system has the ability to expedite the screening of multiple gene cassettes in commercial strains in a single transformation step, and without any preconceived knowledge of the strain’s genetics or target site sequence.
This technology is especially useful in uncharacterized commercial yeast strains with complex genetics and ploidy. For example, users can potentially screen thousands of commercial strains to determine if they are amenable to metabolic engineering, and tolerant of the end product.
In theory, the piggyBac transposase can be further optimized to stably integrate high copy numbers of the transgene into the genome. The system can also accommodate other complex metabolic pathways and even be configured for other fungal species as well.
Overall, the piggyBac system has an enormous cargo-carrying capacity (over 200kb) and can quickly validate a strain for its potential in the biosynthesis of highly valuable metabolites. This type of production can now be accomplished in a more cost-effective and efficient manner with the implementation of the piggyBac system.
Learn More About piggyBac, Cas-CLOVER, And Demeetra
piggyBac can be paired with Demeetra’s Cas-CLOVER targeted nuclease “the clean alternative to CRISPR/Cas9” for knockouts, knock-ins and base-pair edits in the user’s strain of interest. For example, to facilitate the use of nutritional markers and eliminate the use of antibiotic selection markers in potential commercial strains.