BPI West 2024 Poster Presentation: piggyBac Transposase and Cas-CLOVER: CRISPR-based Innovations Enable Rapid Discovery of Novel Hot Spots in HEK293 Suspension Cell Line Development
Cell factory platforms are used in commercial bioproduction to express therapeutic molecules such as antibodies, vaccines, and biosimilars. Through innovative gene editing tools, cell lines can be tailor-made to stably express proteins at varying levels. The piggyBac transposase integrates transposon cargo randomly into stable and highly expressed sites and has been utilized by many pharmaceutical industry leaders for high yield cell line development. This feature of piggyBac makes it a well-equipped tool for mapping genomic hotspots in bioproduction cell lines. Using a HEK293 suspension cell line, the piggyBac transposase/transposon system was utilized to randomly integrate Gene of Interest. High expressing clones were isolated and analyzed using splinkerette PCR to identify the piggyBac integration sites. Cas-CLOVER nuclease was then used to individually knock-in Gene of Interest into the novel locations in wild-type cells to verify expression levels.
piggyBac Transposase Enables Stable Cell Line Development
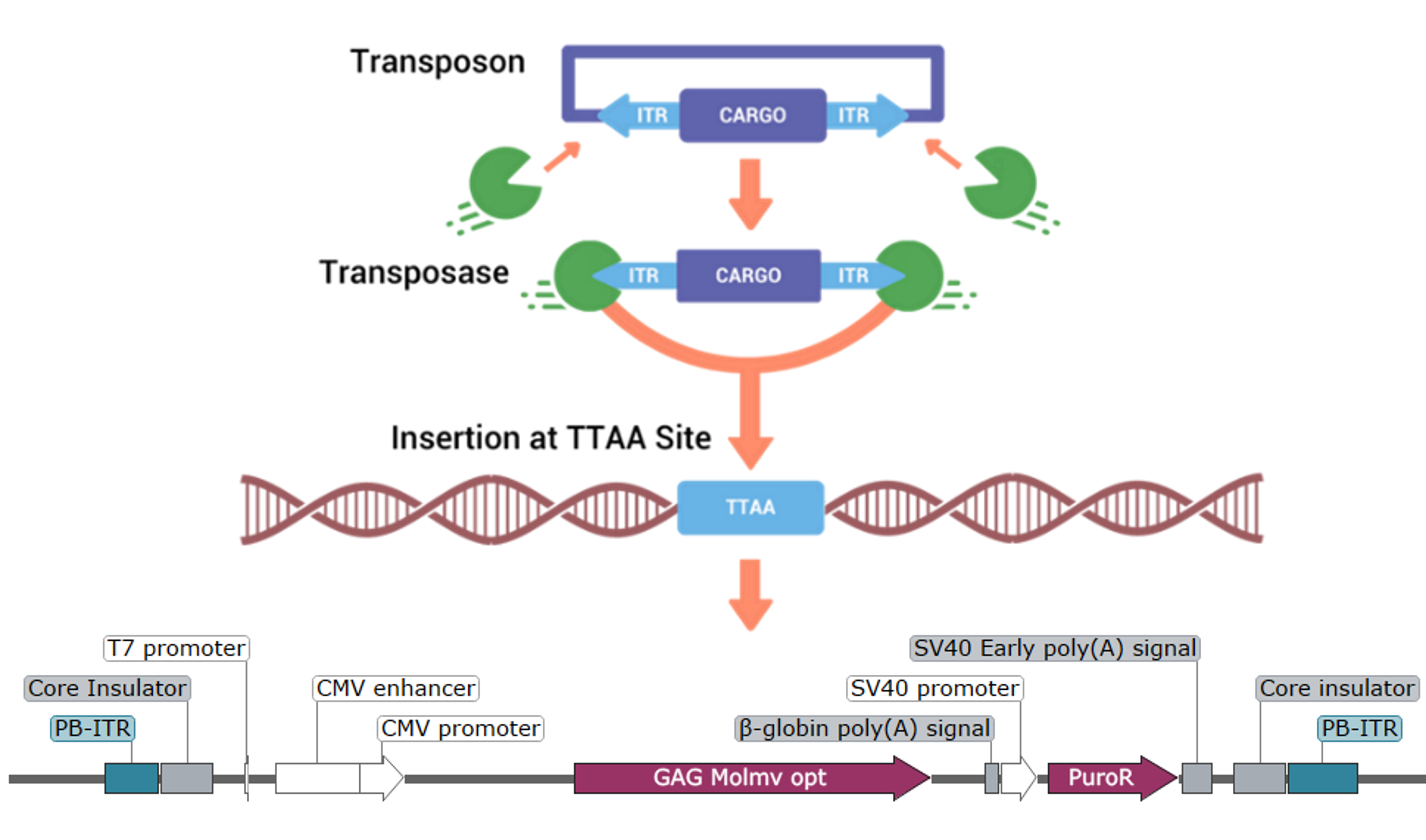
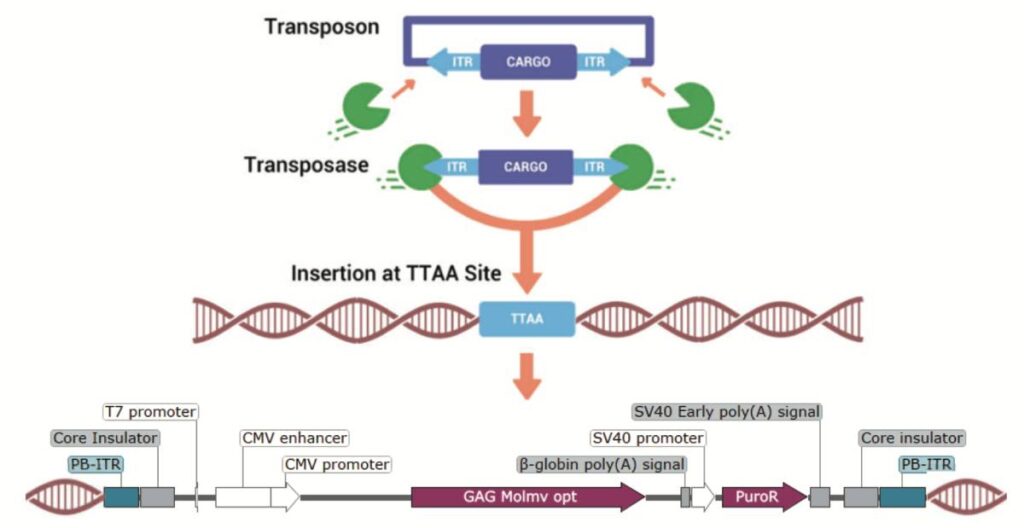
piggyBac Transposase Mechanism: Schematic of the piggyBac transposase mechanism for genomic integration of Gene of Interest with puromycin selection at ‘TTAA’ sites to identify novel regions of interest.
Splinkerette PCR Cell Line Development Workflow
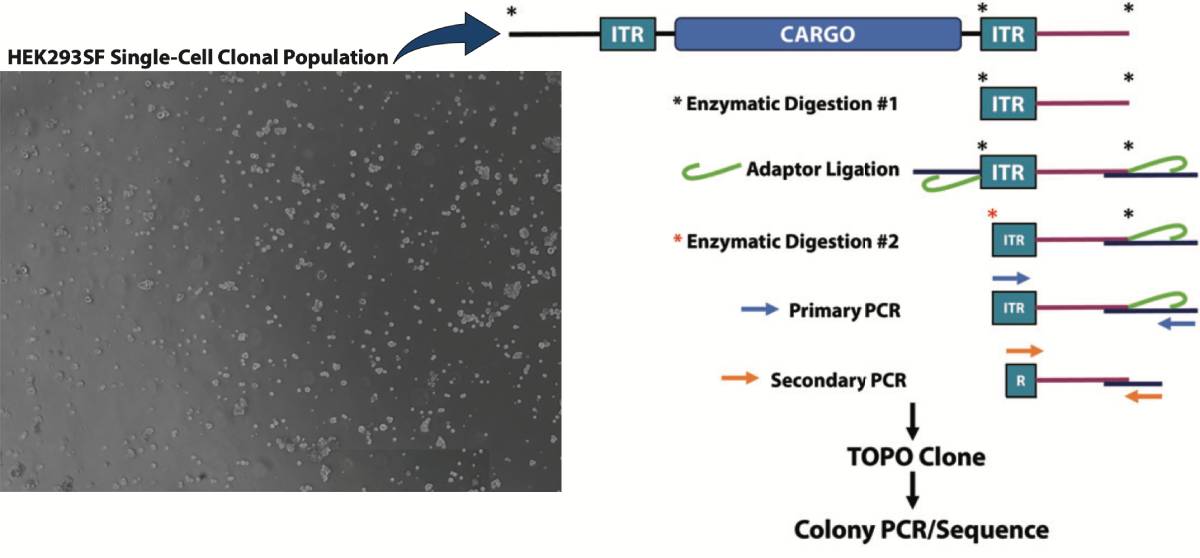
Splinkerette PCR Workflow: Single cell clones were isolated by limited dilution and the clonal populations’ integration sites were analyzed via Splinkerette PCR to identify potential novel sites for variable protein expression. Splinkerette PCR utilizes specific adaptors that form a hairpin secondary structure which serves to also increase specificity.
Identification of Novel Bioprocessing Expression Sites
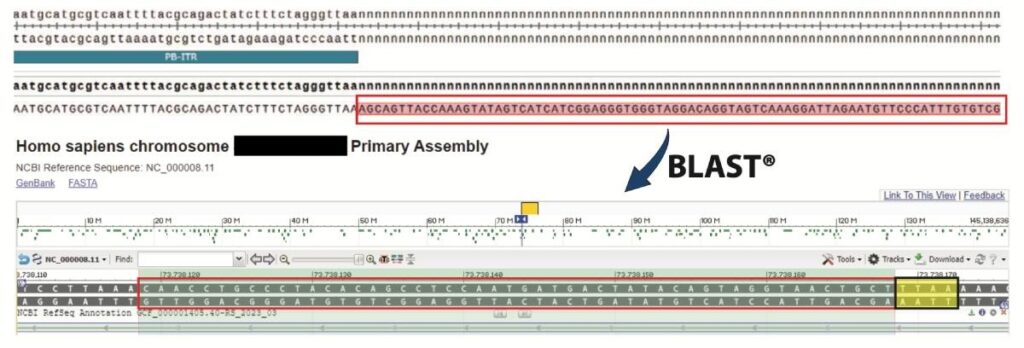
Colony PCR and Sanger sequencing to identify integration site(s): Colony PCRs were performed, product purified, and sequenced. Sequence alignments were made against a custom vector map in SnapGene. 40-60bp nucleotide sequences that flanked the piggyBac ITRs were then used to BLAST the Human genome to identify chromosomal integration sites. The example shown in Figure 3 is an intragenic integration into STAU2 of Chromosome X. The yellow highlighted box depicts the ‘TTAA’ integration site.
Protein Expression Profiles
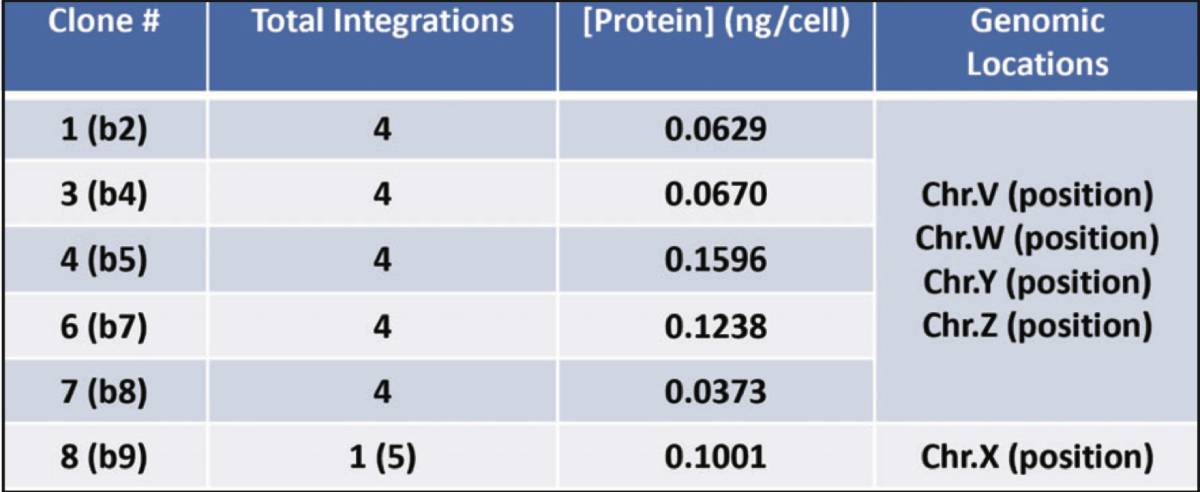
Secreted Gene of Interest protein levels of down-selected piggyBac clones: Several clones were down-selected based on growth and initial Gene of Interest expression. This table represents each clones’ respective Gene of Interest expression profile and the corresponding genomic integration site(s). From the limited dilution, there are several clones with the same integration sites which means that the down-selected pool contained a dominant population of a certain gene-edited cell.
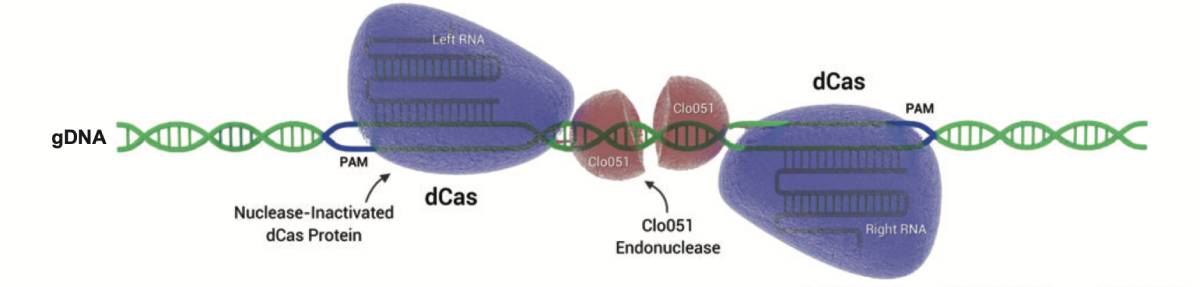
Cas-CLOVER: a dimeric alternative to CRISPR/Cas9
Cas-CLOVER Dimeric Targeted Nuclease and gRNA Pair Efficiencies: Catalytically inactive dCas9 acts as an CRISPR based system to position Clo051 endonuclease, an obligate dimer, into position to cut genomic DNA.
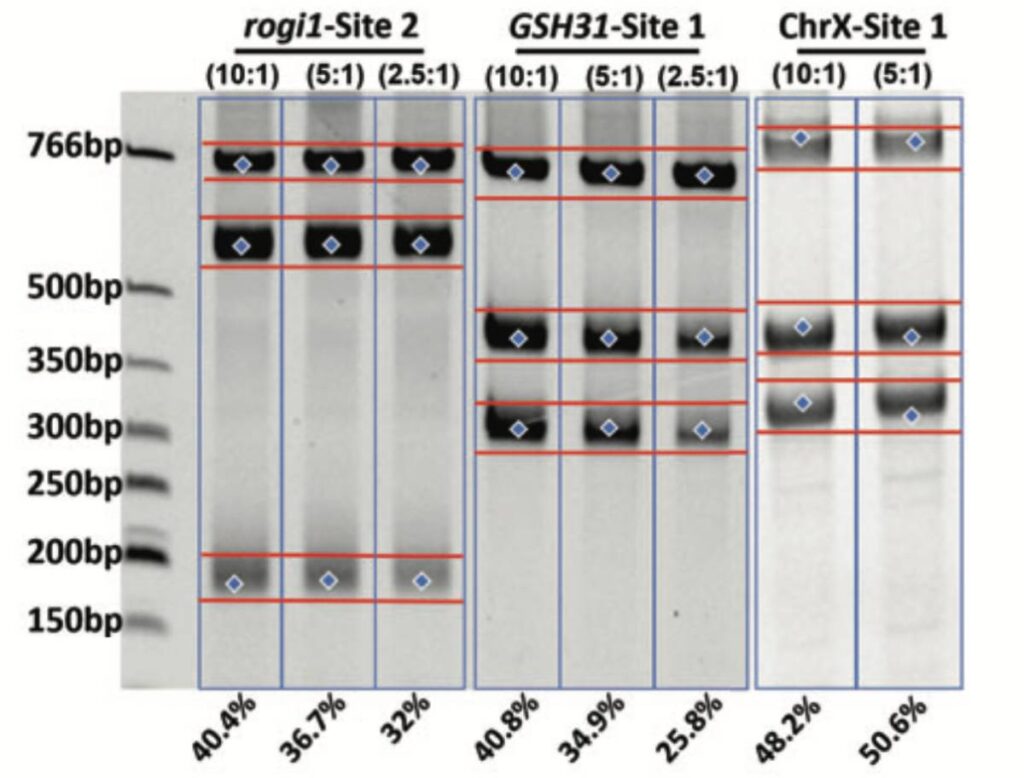
With a flexible spacer length, designing gRNA pairs is simple and effective. Cas-CLOVER mRNA was used in combination with gRNA pairs at varying ratios to achieve >40% cutting efficiencies at three different sites in HEK293SF cells by NEON electroporation.
Cas-CLOVER Mediated Knock-In Verification
Pools under antibiotic selection were assessed for the presence of Gene of Interest knock-in construct via PCR. Amplicons were designed specifically to the Gene of Interest construct and to the flanking 3’ region (2221bp) or 5’ region (2622bp) outside of the 500bp homology arms in the knock-in construct. No PCR product was amplified in the parental HEK293SF cell line as a negative control. Throughout puromycin selection, the amplicon intensity increased as the positive gene-edited cells were enriched.
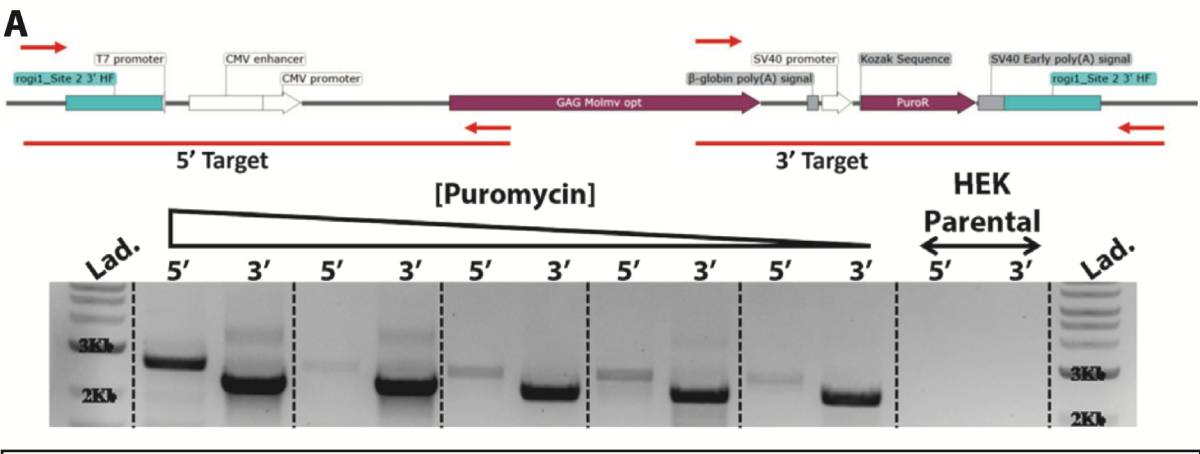

PCR products were isolated and sequenced for verification of correct targeted integration.
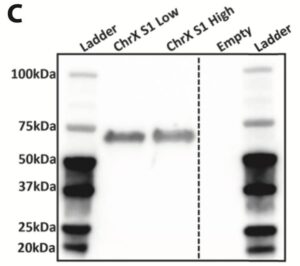
Secreted Gene of Interest protein expression was assessed by western blotting of 30ul of the enriched pool supernatant. An anti-GAG rat mAb and a goat anti-rat HRP-conjugate were used for detection of GAG protein.
Conclusions & Future Research
The technologies forementioned here identified novel hot spots in the genome which surpassed protein expression levels known at genomic safe harbor sites, rogi1 and GSH31. As a result, highlighting the importance and utility of piggyBac transposase to produce innovative cell platforms engineered to consistently produce therapeutic molecules at varying expression levels. Not only can this method be applied to cell lines commonly used in bioproduction, such as CHO and HEK293s, but it can also be used to identify hot spots in novel production lines as well as cell lines used for cell and gene therapy development.
Authors:
- Corey Brizzee, Tyler Kinder, and Jack Crawford from Demeetra AgBio, Lexington, Kentucky
- Jasminka Bozic, Catalina Soare, and Anne-Catherine Fluckiger from 12VBI Vaccines, Ottawa Ontario, Canada
- Daniel Machado and Valeriya Steffey 3Hera BioLabs, Lexington, Kentucky
