While CRISPR based technologies such as Cas-CLOVER can cut DNA with targeted precision, the insertion of very large transgenes or seamless removal of genes, particularly in uncharacterized genomes, necessitates additional tools. The Super piggyBac transposase system employs a transposase/transposon mechanism to introduce or excise genes of varying sizes into host DNA. Furthermore, piggyBac transposase facilitates scarless excision of transgenes for selection marker recycling or phenotype reversion.
How Does piggyBac Work?
PiggyBac is a TTAA-specific transposon, meaning it will only bind to a genetic sequence that has an open reading frame starting with a 4bp “TTAA” sequence. The piggyBac mechanism was originally discovered in Trichoplusia ni, or the cabbage looper moth, but has been used as a mutagenesis and stable cell line development tool in many other eukaryotic organisms, including CHO cells, plants and yeast. As a tool for insertional mutagenesis, piggyBac is able to incorporate mutations into an organism’s genome, allowing for phenotype screening and modification of difficult to edit cells and organisms, especially if homologous recombination (HR) is not efficient.
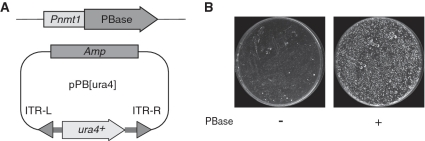
The schematic illustrates piggyBac binary transposition. The pDUAL-PBase plasmid, integrated into the S. pombe genome, expresses PBase under the Pnmt1 promoter. The donor plasmid pPB[ura4] contains PB transposon ITRs with a ura4+ selection marker, lacking transposase (left). The plates display Ura+ transformants of pPB[ura4]. Cells expressing PBase formed more colonies on uracil-free media compared to those without PBase.
Schizosaccharomyces Pombe And Its Significance As A Model Organism
The species of fission yeast, Schizosaccharomyces pombe, has an incredibly simple genome, but unlike S. cerevisiae, it does not have robust genetic tools. Interestingly, S. pombe has a native retrotransposon, Tf1. Although tested for directed mutations, it showed a strong insertional bias against ORFs, meaning it didn't disrupt genes. Because of this, S. pombe is a model that scientists have chosen to integrate with the piggyBac transposase technology. By using the piggyBac transposition tool in S. pombe, researchers were able to perform genetic screens to identify the phenotypes caused by the mutations.
Why Use piggyBac For Mutagenesis And Genetic Screens?
Using Demeetra's piggyBac transposon system for genetic screens in yeast offers several advantages. PiggyBac mutations are easily identifiable and exhibit minimal insertional bias, targeting TTAA sequences. Moreover, piggyBac can remove inserted sequences without leaving any genetic material behind, enabling phenotype observation and confirmation of gene involvement if the original S. pombe phenotype is restored after mutation removal.
References
Li, J., Zhang, J. M., Li, X., Suo, F., Zhang, M. J., Hou, W., Han, J., & Du, L. L. (2011). A piggyBac transposon-based mutagenesis system for the fission yeast Schizosaccharomyces pombe. Nucleic acids research, 39(6), e40. https://doi.org/10.1093/nar/gkq1358
