Characteristics such as robust growth rates, scalability of suspension cultures, and consistent cloning efficiency have established Chinese hamster ovary (CHO) cells as the preeminent mammalian cell line for bioprocessing applications. Nonetheless, the optimization of recombinant CHO cell cultures for the production of biologics, including antibodies, remains a significant challenge.
DHFR vs GS Selection Strategy Overview
The dihydrofolate reductase (DHFR) enzyme facilitates the conversion of folate into tetrahydrofolate, a vital precursor for the de novo synthesis of purines and pyrimidines, which is essential for cellular replication. In the context of protein expression systems utilizing CHO cells researchers introduce recombinant DNA that incorporates the gene of interest in linked to the DHFR gene. The selection of CHO cells that produce the desired protein employs the methotrexate (MTX) selection system. MTX, a folate analog, competitively inhibits DHFR, consequently impeding the synthesis of tetrahydrofolate required for the de novo synthesis of purines and pyrimidines. CHO cells undergo cultivation under progressively increasing concentrations of MTX. This selective pressure favors CHO cells that have multiple copies of the DHFR gene along with the gene of interest, thereby selecting for high expression1.
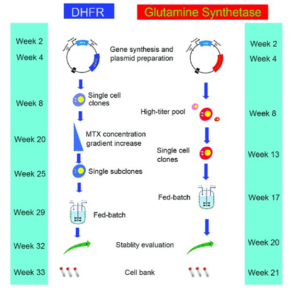
DHFR and GS selection with estimated timelines2.
Glutamine Synthetase (GS) is essential for cell proliferation as it produces of glutamine, enabling cell to replication. Selection based on GS employs the co-expression of the GS gene alongside the gene of interest as a selective marker for identifying CHO clones with high GS expression levels. Given that CHO cells inherently synthesize glutamine, a GS inhibitor, Methionine Sulfoximine (MSX), is required during the selection process. This GS-based selection methodology represents an advancement over the Dihydrofolate Reductase (DHFR) system, primarily because it necessitates a reduced selection intensity, thereby potentially shortening the developmental timelines.
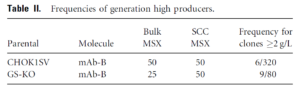
One key flaw to MSX selection is CHO cells can maintain a low endogenous glutamine level and often survive the MSX selection. This decreases the GS/MSX selection stringency. Therefore, creating CHO cell lines with a complete GS knockout as an expression system would be ideal. Researchers at Eli Lilly used zinc finger nuclease (ZFN) technology to knockout GS with an efficiency of 2%, very low by contemporary standards. Nevertheless, GS CHO knockout cells exhibited more stringent selection, with 6-fold increase in high producers3.
The Potential of Next Generation CRISPR-Based Tools for Therapeutic Bioprocessing
Using CRISPR and ZFNs to improve CHO cells through GS knockout is just one example of how gene editing is changing bioprocessing. Demeetra's Cas-CLOVER differs from Cas9 as it utilizes Clo51, a dimeric endonuclease that is only active upon dimerization. This requires two guide RNAs (gRNAs) to bring nuclease inactivated Cas9 proteins to the site where cutting is desired, thereby bringing the dimers together. This dimerization requirement results in higher accuracy for Cas-CLOVER than regular CRISPR-Cas9, while maintaining very high efficiencies. Learn more about how we used Cas-CLOVER to create a functional knockout of GS, the CleanCut GS CHO Cell Line.
Cas-CLOVER not only surpasses the constraints inherent in earlier gene-editing methodologies such as ZFNs but also distinguishes itself from the fundamental CRISPR/Cas9 intellectual property (IP), facilitating a more unambiguous path to operational freedom. We invite researchers to investigate the capabilities of Cas-CLOVER to augment their gene editing initiatives. By partnering with Demeetra, clients gain not merely access to cutting-edge tools and IP licenses but also enter into a collaborative relationship that encompasses exclusive protocols, specialized knowledge, and direct technical assistance from our staff, whose expertise is validated through peer-reviewed publications. We encourage interested parties to contact Demeetra to discover how our bespoke gene editing solutions can be adapted to fulfill specific research and development objectives.
- Ng. (2012) Mol. Methods. Biol
- Yang (2022) Frontiers Bioeng. and Biotech.
- Fan (2012) Biotech and Bioeng.