Bioprocessing International 2024 Poster: Cas-CLOVER-mediated knockout of STAT1: A novel approach to engineer packaging HEK293 cell lines used for rAAV production
To address the limitations of CRISPR-Cas9, including off-target effects and high licensing costs for commercial use, the Cas-CLOVER system, a dimeric gene-editing tool activated by two guide RNAs, was recently developed. Here we evaluated the application of Cas-CLOVER in suspension HEK293 cells used for recombinant adeno-associated virus (rAAV) production by targeting the STAT1 locus, a critical regulator of cell growth that may influence rAAV production yields.
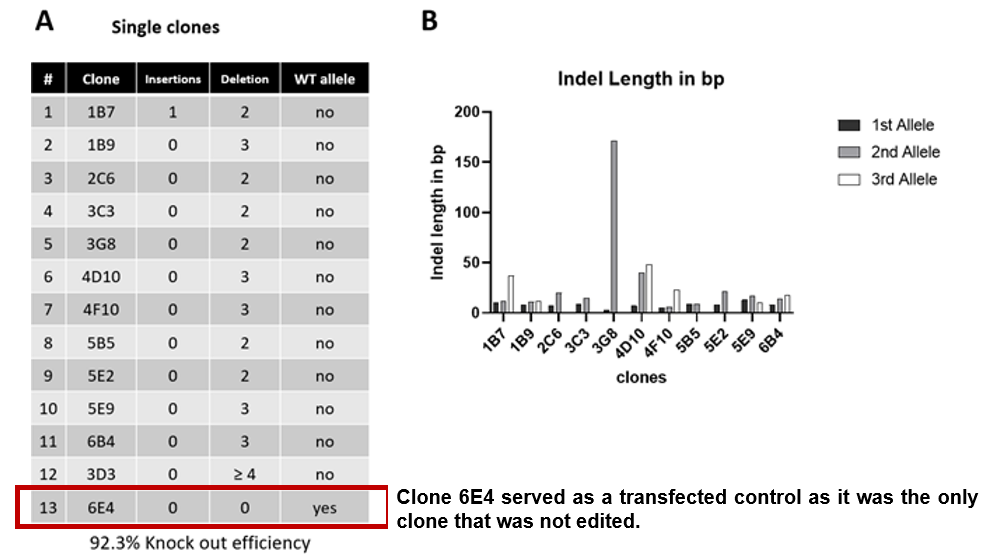
- Cas-CLOVER demonstrated an editing efficiency of 92.3%, consistent with the highest values reported in the literature
- All target loci were edited, with no wild-type sequences detected among the edited clones
- The mutations primarily consisted of larger deletions
- STAT1 protein was completely absent confirming disruptive knockouts (KO)
- Capsid and vector genome titers, and levels of viral capsid and replication proteins were highly variable amongst the KO clones
Cas-CLOVER Mediated STAT1 Knockout in Suspension HEK293 Cells
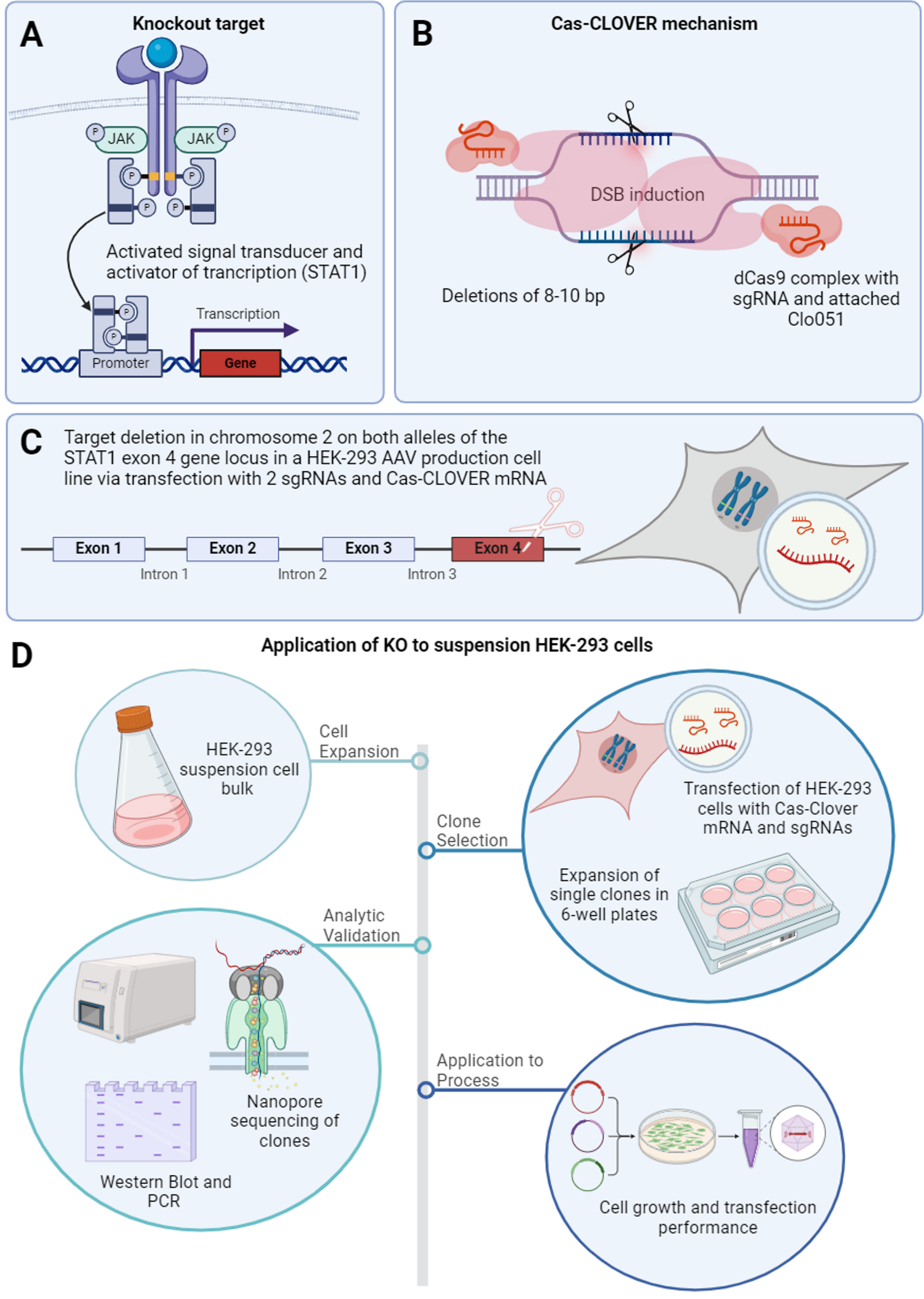
A) STAT1 pathway and knockout target. B) Cas-CLOVER deletion mechanism.
C) Targeted cutting loci.
D) Summary of STAT1-KO application through suspension HEK-293 cell line, clone selection process, analytic performance validation by JESS, ddPCR and nanopore sequencing, and final cell growth and transfection performance of STAT1-KO clones.
Cas-CLOVER Demonstrates 92.3% Gene Editing Efficiency
Analysis of STAT1 Knockout Efficiency in HEK293 Cells: A) Nanopore sequencing demonstrates a 92.3% knockout efficiency across 13 selected clones, featuring a range of deletions. B) Graph depicting the average length of insertions and deletions (indels) in base pairs for various alleles among the analyzed clones.
No Detection of STAT1 Protein Edited Clones
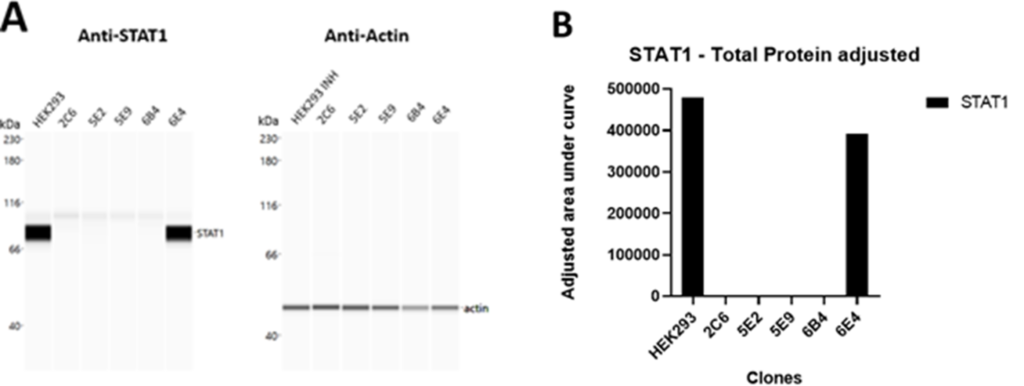
Validation and Characterization of STAT1 Knockout in HEK-293 Cells. A) Western blot analysis reveals absence of STAT1 protein in all KO clones, with HEK293 bulk cells and clone 6E4 (WT allele) serving as controls. Anti-Actin, the positive control protein, was detected in all samples. B) The adjusted area under the curve analysis indicates marginally lower STAT1 levels in clone 6E4 (WT allele) compared to HEK293 bulk cells.
- No STAT1 protein was detectible, regardless of whether the INDELs resulted in a frame shift or in-frame mutation.
- Our data suggests the larger deletions increased the likelihood of achieving a disruptive knockout
Performance of STAT1 KO clones in rAAV virus production
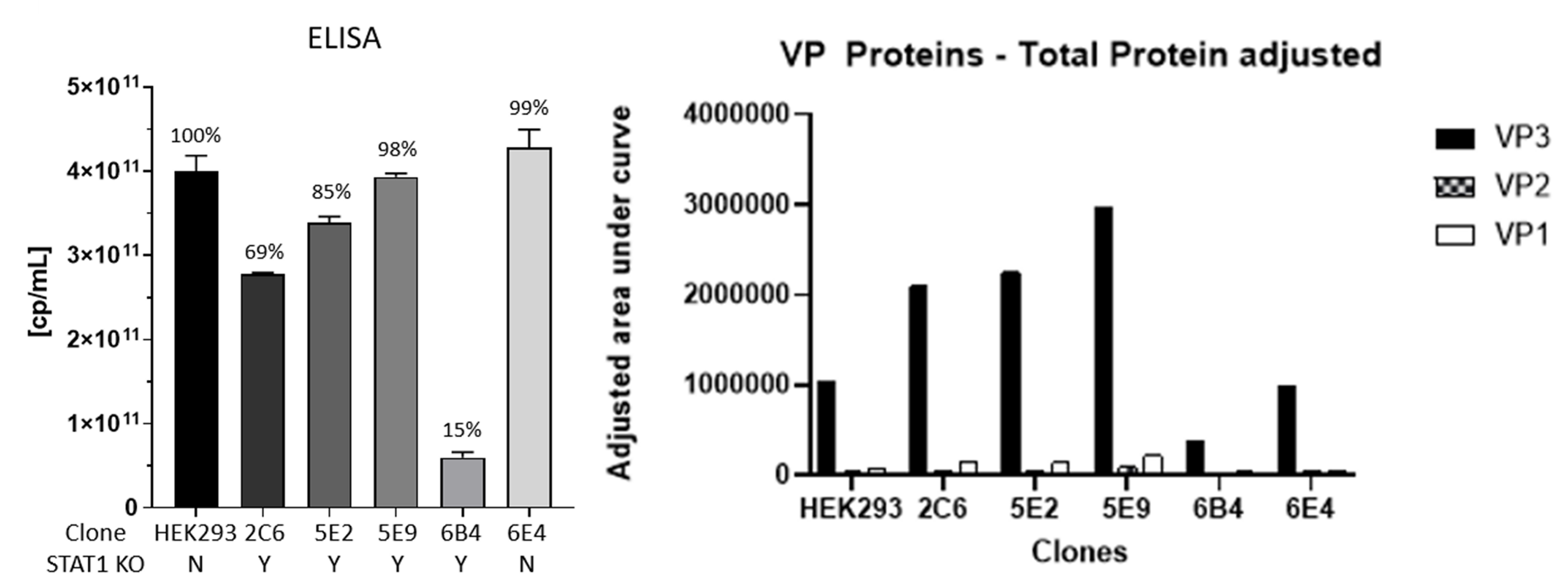
Comparative Analysis of Viral Production and Protein Expression in HEK293 Cells Post STAT1 Knockout. (Left) AAV-specific ELISA capsid/mL titer. (Right) Analysis of viral proteins (VP) 1, 2, and 3.
Conclusions & Opportunities
In summary, this study demonstrates the applicability of the Cas-CLOVER system in an rAAV-producing HEK-293 cell line. The high editing efficiency and minimal off-target effects make Cas-CLOVER a promising alternative to the standard CRISPR/Cas9 system. Although single STAT1 knockouts did not enhance rAAV yield, the potential for engineering cell lines for improved AAV production remains substantial. Leveraging gene-editing tools like Cas-CLOVER, alongside the genetic diversity of HEK-293 cells and high-throughput technologies, could lead to significant yield improvements.
Reference
Andorfer, P., Kahlig, C.-I., Pakusic, D.,Pachlinger, R., John, C., Schrenk, I., Eisenhut, P., Lengler, J.,Innthaler, B., Micutkova, L., Kraus, B., Brizzee, C., Crawford, J.,& Hernandez Bort, J. A. (2024). Cas-CLOVER-mediated knockout of STAT1: A novel approach to engineer packagingHEK-293 cell lines used for rAAV production.
BiotechnologyJournal, 19, e2400415. https://doi.org/10.1002/biot.202400415
