Parthenocarpy, the production of seedless fruit without fertilization, occurs naturally either due to mutations or as a defense against predators. Scientists also exploit it to extend shelf life or for practical uses like large-scale sauce production and customer appeal.
Seedless watermelons from grocery stores are smaller, making them easier to carry and less wasteful for small households. Consumers also appreciate easier clean-up with seedless options.
Gene editing technology efficiently alters plant genomes to obtain desirable traits, whether for practicality, functionality, or aesthetics and convenience.
Target Developmental Genes?
Traditionally, horticulturists breed plants by increasing the likelihood of producing certain genes as is done in the natural selection process. A gene of interest is identified for a specific characteristic and then amplified in the population by breeding those plants with the genetic characteristic of interest.
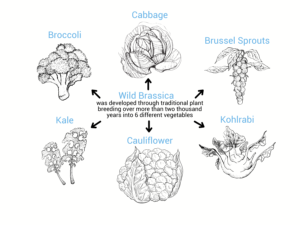
Figure 1: Traditional genetic engineering has done amazing things, but is inefficient and time-consuming
Advancements in genetic modification techniques now allow us to alter organisms with greater precision. It is imperative to comprehend signaling pathways and genetics thoroughly to prevent any unintended alterations. In the context of fruit development, research may target developmental genes and their associated pathways.
The auxin response pathway plays a pivotal role in regulating the transcription of these genes. Targeting this pathway presents an opportunity for the development of gene editing tools aimed at modifying seed production in plants.
Altering Tomato Protein With Gene Editing Technology
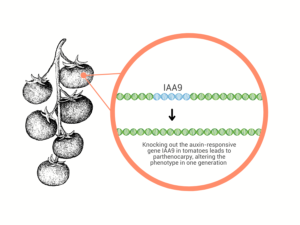
Figure 2: Gene editing technology can introduce desired traits in one generation, unlike traditional breeding
A recent study focused on using CRISPR/Cas9 gene editing technology to alter the auxin-responsive protein, SIAA9, in the tomato (Solanum lycopersicum) plant (1). Aux/IAA9 and auxin response factor 8 (ARF8) are involved in repressing fruit initiation without fertilization (parthenocarpy) in tomatoes. The researchers found that the SlIAA9 knock outs exhibited simple leaves compared to the compound leaves of the wild-types; shown in Figure 3.
Additionally, fruit development was triggered before fertilization in those plants without SlIAA9, giving rise to seedless tomatoes; also shown in Figure 3. Furthermore, the mutations were heritable in subsequent generations meaning it’s a sustainable genetic editing technique (1)
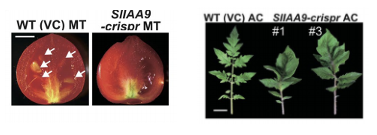
Figure 3: Tomato fruit with seeds (WT) and without (genetically modified fruit). Compound leaves in wild-type vs. simple leaves in the genetically modified leaves.
Novel Tools For Gene Editing In Plants
Previous research in fruit ripening enzymes used gene technology such as RNAi which reduces gene expression at the mRNA level while newer technologies such as CRISPR and Cas-CLOVER completely and permanently silences the gene at the DNA level. Novel gene editing proteins such as, Cas9, transcription activator-like effector nucleases (TALEN), and zinc-finger nucleases (ZFN) target specific genes in a deliberate manner. This is compared to previous tools that randomly insert, delete, or cause a frameshift in the sequence being edited; dramatically altering gene function.
Besides the temporary reduction vs. a permanent solution, RNAi silencing method suffers limitations due to high off-target effects. Off-targets in plants could lead to unwanted changes that reduce quality possibly leading to decreased yield, income, and /or health and regulatory concerns. The Cas-CLOVER gene editing system shown in Figure 4 utilizes a catalytically inactive Cas9 protein fused to the Clo51 nucleus domain which works as monomers recruited by a pair of guide RNAs (gRNA) to introduce targeted mutations. And when both subunits are properly recruited to the target-site, it leads to dimerization and activation of the Clo051 nucleus domain, leading to targeted gene disruptions.
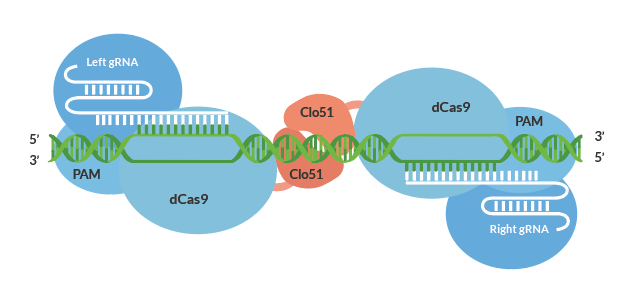
Figure 4: Cas-CLOVER gene editing system
Demeetra’s Cas-CLOVER Gene Editing Systems
Advanced gene editing technologies enable plant biotechnologists with the ability to make targeted knockouts and insertions. This scarless gene editing means no heterologous genes are left in the plant genome, producing a non-GMO crop and simplifying the process. We recognize the impact that gene editing tools have on the future of science, agriculture, and ultimately food preservation. In particular, we offer licenses for our novel gene editing Cas-CLOVER system.
Shown in a previous publication, we were able to validated the activity of Cas-CLOVER in plants by targeted inactivation of the RNA-dependent RNA polymerase 6 (RDR6) gene in tobacco achieving an efficiency of at least 18% in the first target which is significant for tobacco.
Enhancing tomato production is just one example that can benefit from gene editing technology. Our rapidly growing population with threats of climate change continue to be factors that pressure our food and medicine security, increasing the need for more productive agriculture traits and biomanufacturing systems is essential.
If you’d like to learn more about Cas-CLOVER, please contact us to schedule a meeting online.
References
- Ueta, R., Abe, C., Watanabe, T., Sugano, S. S., Ishihara, R., Ezura, H., … Osakabe, K. (2017). Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Scientific Reports, 7(1), 1–8. https://doi.org/10.1038/s41598-017-00501-4
