The CRISPR Cas9 system has revolutionized targeted genome engineering owing to its simplicity in employing guide RNAs. However, this technology is not without its limitations. Notable among these are the predisposition for off-target mutagenesis, stringent licensing restrictions with lack of clear freedom to operate (FTO), and economically burdensome license terms.
To address the limitations of the single-guide RNA (sgRNA) CRISPR Cas9 editing method, researchers at Demeetra initiated a study to evaluate the potential of its Cas-CLOVER technology as an alternative for agricultural gene editing. This study aimed to validate Cas-CLOVER's nucleic acid cleavage efficiency in tobacco as a research model, suggesting it as a precise and cost-effective genetic editing tool for agricultural biotechnology.
How Cas-CLOVER Works And How It Differs From Single-Guided CRISPR Cas9 Gene Editing
Cas-CLOVER is a gene editing system similar to CRISPR/Cas9 but offers unique advantages.
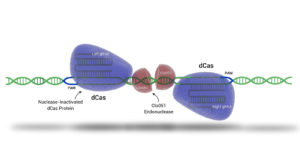
While CRISPR/Cas9 uses a single guide RNA and Cas9 to cut DNA, Cas-CLOVER uses two guide RNAs and a deactivated Cas9 for binding without cutting. The cutting is done by Clo051, which enhances precision and reduces off-target mutations. Additionally, Cas-CLOVER creates larger 4-base pair deletions, leading to more complete gene knockouts and better efficiency for gene insertions.
These unique features of Cas-CLOVER improve its editing accuracy and establish its own intellectual property.
Demonstration of Cas-CLOVER Cutting Efficiency In Plants
Cas-CLOVER has shown effective targeted mutagenesis in mammalian cells, and we aimed to apply it in plants for crop trait discovery and development. We focused on the phytoene desaturase (PDS) gene in tobacco due to its white phenotype. Screening green, pale, and white shoots showed indel efficiencies. The fully white phenotype indicated all four alleles were knocked out with Cas-CLOVER. After optimizing protocols, we increased white shoot numbers and cutting efficiencies to over 90%, as shown in the representative plate below.
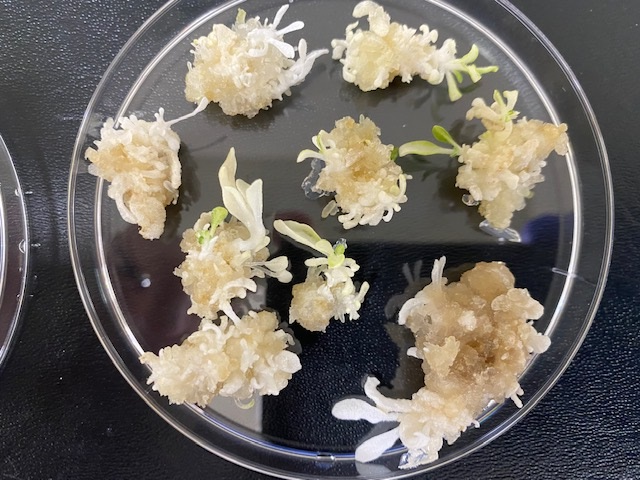
Phenotype of successfully edited tobacco shoots
Genetic modifications introduced using the Cas-CLOVER gene editing system were evaluated through a comprehensive screening approach. This process involved the amplification of target regions via polymerase chain reaction (PCR), followed by Sanger sequencing to identify specific nucleotide changes. Additionally, the efficiency and outcomes of the editing events were assessed using the Synthego Inference of CRISPR Edits (ICE) analysis, which quantifies insertion and deletion (indel) events at the targeted genomic loci. The ICE system provides detailed indel percentages, offering a robust and quantitative evaluation of editing outcomes.
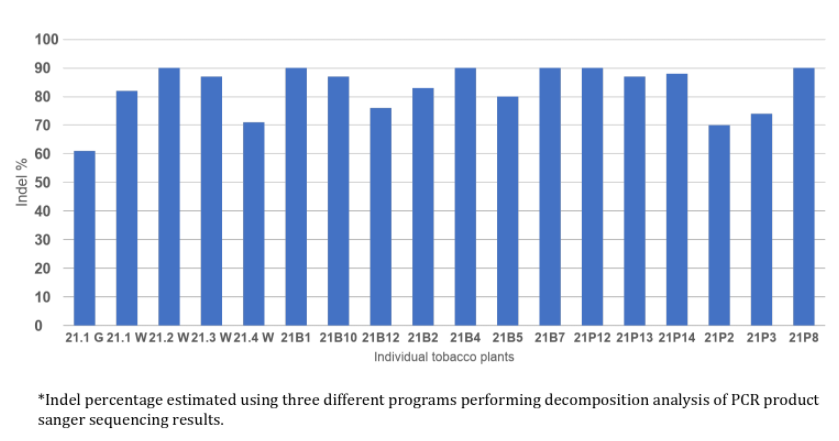
Estimated indel frequencies of individual plant subjects
Improved Downstream Processes For Cas-CLOVER Editing Through The plant T1 Generation
Eight (8) T0 edited plants were grown to the flowering stage, the plants were self-pollinated, and seeds were harvested. Following seed germination, seedlings were transferred to magenta boxes and grown large enough to check for edit stability in this T1 generation. Unpurified PCR products were screened using an optimum resolution on an automated DNA analysis machine, QIAxcel (<500bp size).
- Cas-CLOVER produces larger indels than other technologies
- The large indels are rapidly and easily detected by peak size & translated into a gel image
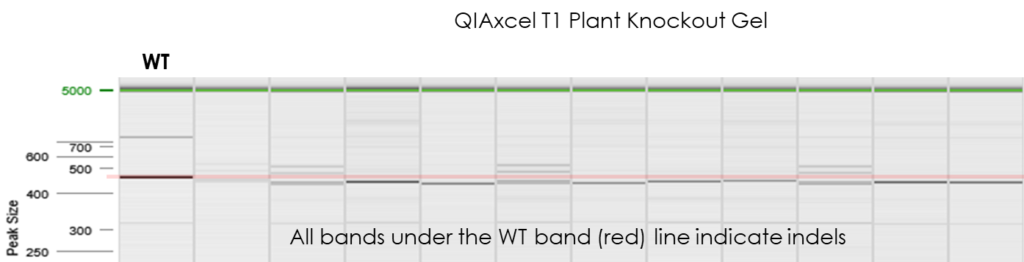
QIAxcel results demonstrating very high stability in the edited genome.
T1 indels were then double checked using Synthego ICE. 14 of 16 samples sequenced and analyzed demonstrated 100% indel percentage. Two samples showed indel and WT sequences by both QIAxcel and ICE. Two other samples with WT peaks by QIAxcel revealed edited but smaller deletions by ICE not picked up in the QIAxcel screen.
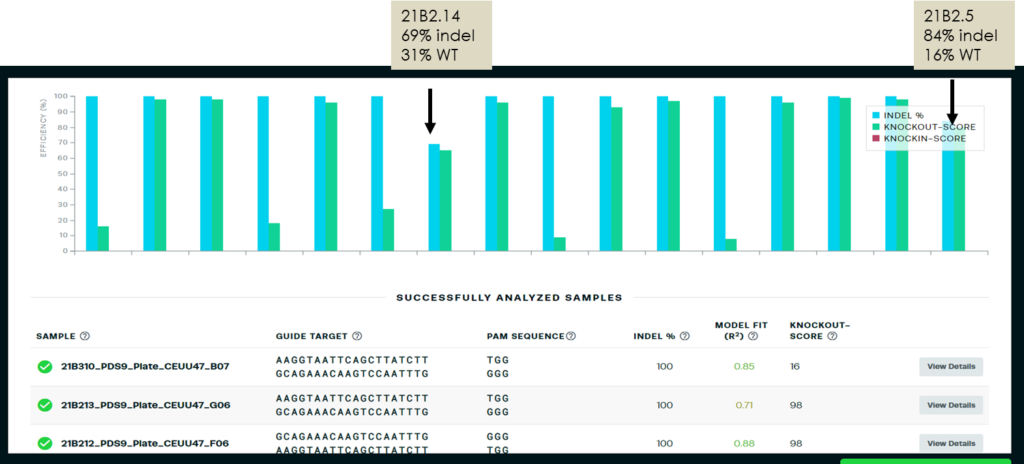
Synthego ICE results reflecting 100% indel for 14 of the 16 specimens and partial indel for the other 2
Demeetra’s Approach To Cas-CLOVER Licensing
This pioneering work conducted by Demeetra in refining the Cas-CLOVER technology presents a significant leap forward in the field of agricultural biotechnology, particularly in the realm of crop improvement. By addressing the limitations of the traditional CRISPR Cas9 system, such as off-target effects and licensing, Cas-CLOVER emerges as a highly precise, cost-effective, and efficient alternative for targeted genome engineering. Its unique dimeric gene editing mechanism, leveraging dual guide RNAs and the proprietary Clo051 endonuclease, has demonstrated substantial promise in plant model organism modifications, showcasing both high mutagenesis efficiency and edit stability through generations. This proof of concept establishes the versatile applicability of Cas-CLOVER across various plant species, offering new horizons for trait discovery, genetic improvement, and ultimately, the enhancement of crop resilience and productivity.
References
- Norman, D., Rector, K., Tateno, M., & Crawford, J. (2020). Targeted editing of Tobacco with Cas-CLOVER™: the clean alternative to CRISPR/Cas9 for plant genome editing. Plant Biology 2020. Demeetra AgBio, Lexington, KY
