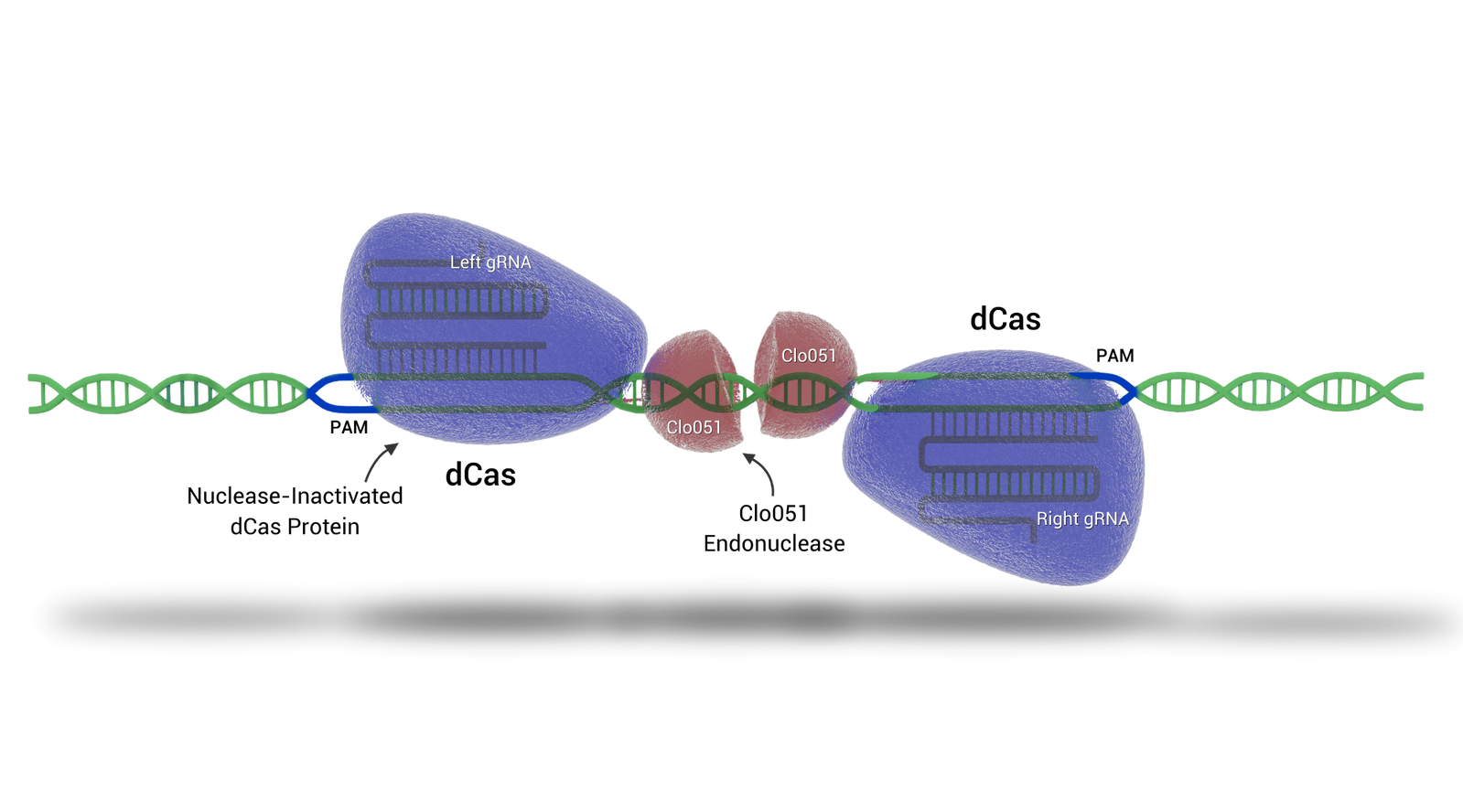
Cas-CLOVER
Cas-CLOVER 'the clean alternative to CRISPR/Cas9' introduces targeted double-strand breaks in genomic DNA, enabling knockouts and site directed knock-ins with high precision.
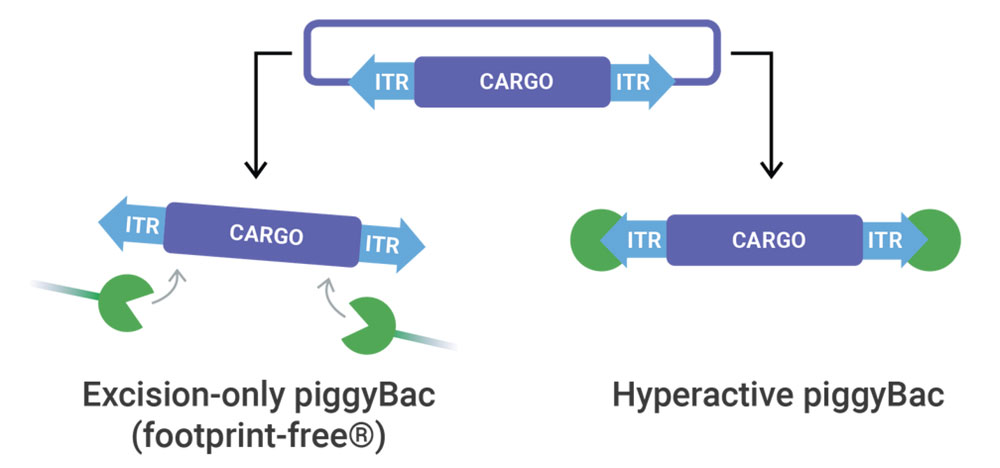
piggyBac Transposase
At elevated efficiencies, piggyBac transposase delivers any sized genetic cargo for stable and high expression. When Footprint-Free gene editing is required, excision-only piggyBac seamlessly removes genetic cargo.
Validated In Mammalian Cells, Yeast, and Plants
We optimize gene editing platforms for broad commercial applications and organisms.
Pharmaceutical Bioprocessing
Bioprocessing and cell line engineering for therapeutic biomanufacturing
Synthetic Biotechnology
Microbial strain engineering to produce therapeutics, industrial enzymes, compounds or biofuels
Agriculture Biotechnology
Targeted editing in plants for transgene-free trait discovery and development
Trusted By Leading Biotechnology Organizations








As Seen In Top Tier Publications
Our gene editing technology has been featured in leading publications.



We provide commercial gene editing licenses and exceptional know-how transfer
Contact us to learn more about our independent gene editing technology IP, optimized reagents and internal expertise to help guide smooth adoption.
