Despite the availability of established mammalian cell expression systems, the production processes for biotherapeutic proteins, antibodies, viral vectors, and vaccines remain notably laborious and financially demanding. A significant factor contributing to these high costs includes inefficiencies inherent in the platform cell lines, such as CHO (Chinese Hamster Ovary) and HEK293 (Human Embryonic Kidney 293) cells.
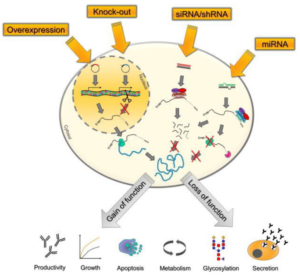
The Cas-CLOVER gene editing technology presents a universal solution, demonstrating efficacy across many cell types and suitability for a diverse array of projects, including targeted knock-ins, broad platform enhancements such as high-yield clonal selection strategies employing glutamine synthetase (GS), among others.
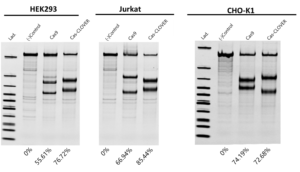
In three separate cell lines at different genomic targets, we found Cas-CLOVER’s activity significantly outperformed or equal to Cas9 as measured by Indel frequency in the T7 EnGen cleavage assay.
The Cas-CLOVER system leverages an innovative hybrid gene editing approach that combines a nuclease-inactivated Cas protein with the Clo051 endonuclease. The mechanism of DNA cleavage is reliant on the dimerization of Clo051, which is an "obligate dimer." This process enables precise targeted gene knockout or knock-in events through either non-homologous end-joining (NHEJ) or homologous directed repair (HDR), offering greater on-target accuracy compared to other gene editing methodologies. Cas-CLOVER's differentiated composition and method carves out a separate intellectual property domain, enabling autonomous operational capability.
The Power of Clean, Targeted Knockouts
The application of Cas-CLOVER technology for gene knockout offers significant advancements in bioprocessing capabilities, affording researchers the opportunity to explore an expansive potential in cell line development and biotherapeutic enhancements. Specifically, in glycoengineering, the targeted disruption of the fucosyltransferase gene (FUT8) facilitates the production of afucosylated monoclonal antibodies (mAbs), thereby enabling the generation of novel and enhanced biotherapeutic entities. Additionally, the knockout of certain host cell proteins, including annexin A2, matripase-1, lipoprotein lipase, and cathepsin D, can mitigate the degradation of drug substances. This intervention not only conserves cellular resources but also augments the productivity and purity of antibodies. Beyond traditional growth selection (GS) null platforms, the elimination of Fam60A has been shown to improve the selection process for clones that exhibit elevated stability in protein production. Enhancements in viral bioproduction have also been achieved through the knockout of proapoptotic proteins such as Bak and Bax, resulting in increased cellular viability and consequently, higher viral titers. This multifaceted approach underscores the transformative potential of gene knockout strategies in the optimization of bioprocessing outcomes.
Knock-In Genes of Interest Using Cas-CLOVER
Unstable and variable transgene expression as a result of random plasmid integration, has historically impeded the efficiency of cell line development (CLD). Such variability frequently introduces delays into projects, thereby causing bottlenecks in the CLD pipeline. To address this prevalent challenge within upstream processing, the Cas-CLOVER system offers a solution through its capacity for precise, site-specific integration of single or multiple copies of the gene of interest.
HEK293 cells are an important platform for vaccine and gene therapy biomanufacturing. As a proof of concept, we used Cas-CLOVER to knock-in a large 10kb insert containing sequences that encode the proteins of interest at a well-characterized genomic safe harbor site.

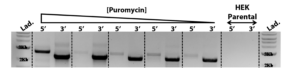
The construct containing the gene of interest, selection elements and the flanking arms of homology as well as the 3’ and 5’ genomic flanking regions are shown with primer sites for PCR amplification.
Pools under selection were assessed for the presence of the knock-in construct via PCR. Amplicons were designed specifically to the gene of interest and to the flanking 3’ region (2221bp) or 5’ region (2622bp) outside of the 500bp homology arms in the knock-in construct. No PCR product was amplified in the parental cell line as a negative control. Throughout puromycin selection, the amplicon intensity increased as the positive gene-edited cells were enriched.
PCR products were isolated and sequenced for verification of correct targeted integration. Secreted protein expression was assessed by western blotting of the enriched pool supernatant.
By enabling precise gene knockout and knock-in modifications, Cas-CLOVER can provide a transformative approach for enhancing cell line development processes. Whether through the production of afucosylated monoclonal antibodies, the improvement of cellular viability for increased viral titers, or the precise integration of genes, Cas-CLOVER's broad applicability spans the requirements for comprehensive platform enhancements. We invite researchers and biotechnology professionals to explore the potential of Cas-CLOVER and to collaborate with Demeetra in leveraging this versatile technology. Through its peer-review published staff, Demeetra offers access to a wealth of expert knowledge, advanced protocols, and dedicated technical support, ensuring that our partners are well-equipped to harness the full capabilities of gene editing in cell line development.