Learning Hub
Presentation
Poster Presentation: piggyBac® Transposase and Cas-CLOVER™ Targeted Nuclease Enable Rapid Discovery of Novel Hot Spots in HEK293 Suspension Cells
Read More
Events & News
Demeetra Cas-CLOVER Research Selected as a MDPI Journal Editor’s Choice Article
Read More
Blog
Agriculture BiotechnologyBioproductCell BioprocessingSynthetic Biotechnology
Demeetra’s Guide to CRISPR and Gene Editing Licensing
Read More
Presentation
Agriculture Biotechnology
Q&A Podcast: Cas-CLOVER Capable of Meeting Crop Improvement Challenges
Read More
Presentation
Synthetic Biotechnology
Q&A Podcast: The Possibilities of Cas-CLOVER and piggyBac
Read More
Peer-Reviewed Publications
A piggyBac-mediated transgenesis system for the temporary expression of CRISPR/Cas9 in rice
Read More
Peer-Reviewed Publications
Cas-CLOVER is a novel high-fidelity nuclease for safe and robust generation of TSCM-enriched allogeneic CAR-T cells
Read More
Presentation
Cell BioprocessingSynthetic Biotechnology
Protein Engineered Cas-CLOVER Yields 99% Gene Editing Efficiency and Ultra-Low Toxicity
Read More
Cell Bioprocessing
Cas-CLOVER: The Proven Alternative To CRISPR/Cas9 For Pharmaceutical Bioprocessing
Read More

Presentation
Cell BioprocessingSynthetic Biotechnology
Q&A Podcast: Advanced Alternatives to CRISPR/Cas9
Read More
Agriculture BiotechnologyCell BioprocessingSynthetic Biotechnology
Gene Editing May Soon Be Rolling in Clover
Read More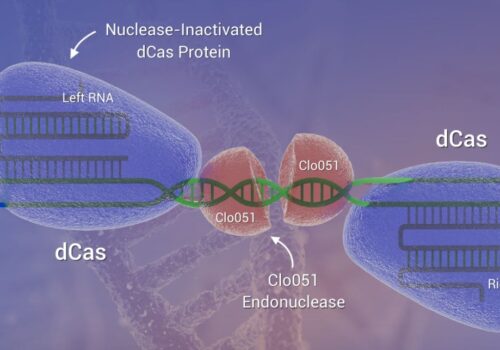
Agriculture BiotechnologyCell BioprocessingSynthetic Biotechnology
The Greatest Gene Editing Technology on the Planet?
Read More
Blog
Synthetic Biotechnology
Accelerating Cultured Meat and Alternative Food Protein Production: The Power of Cell Engineering and Gene Editing
Read More
Agriculture BiotechnologyCell BioprocessingSynthetic Biotechnology
Designing gRNA User Guide
Read More
Blog
Agriculture Biotechnology
Gene Editing in Plants is Stable, Efficient, and Reliable With Cas-CLOVER
Read More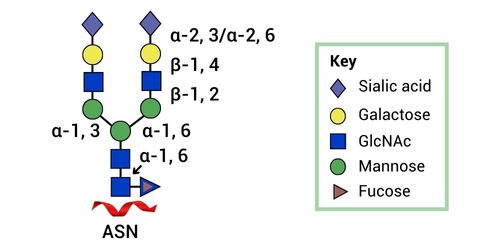
Blog
Cell Bioprocessing
Glycoengineering Strategies For Improved Biologics Manufacturing In Mammalian Cells Using Cas-CLOVER
Read More
Blog
Cell Bioprocessing
Unlocking the Future of Gene Therapy, rAAV And Vaccine Bioprocessing: Advanced Cas-CLOVER Gene Editing in HEK293 Cells
Read More
Blog
Agriculture BiotechnologyBioproductCell BioprocessingSynthetic Biotechnology
Precision at the Forefront: Cas-CLOVER’s Leap Over CRISPR/Cas9 in Target-Specific Gene Editing
Read More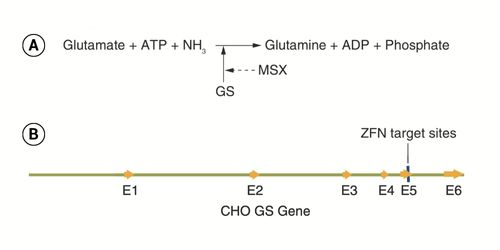
Blog
Cell Bioprocessing
DHFR/MTX and GS/MSX Selection in CHO Cells for Biologics Manufacturing
Read More
Presentation
Cell Bioprocessing
Demeetra/Elanco Webinar: Cas-CLOVER Update – Cell Line Development And Engineering
Read More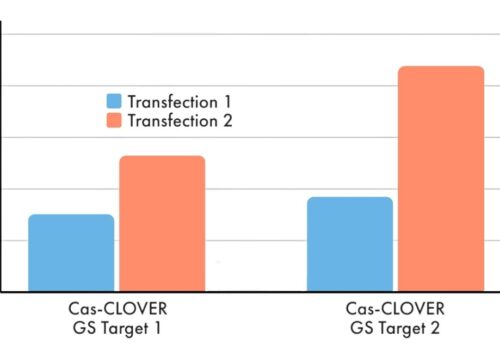
Blog
Agriculture BiotechnologySynthetic Biotechnology
Unlocking Genetic Potential: Target Flexibility and Larger Deletions With Demeetra’s Cas-CLOVER Technology
Read More

Blog
Agriculture Biotechnology
Taking the Potato From Genetically Engineered to Seamless Trait Development with Cas-CLOVER and piggyBac Technologies
Read More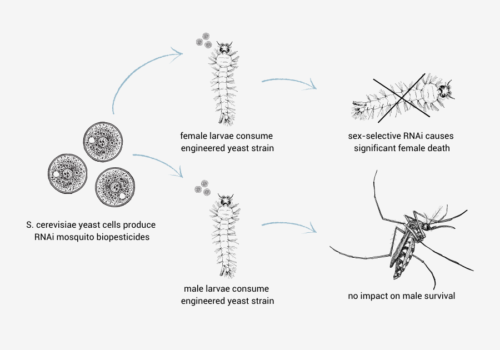
Blog
Synthetic Biotechnology
RNAi Biopesticide Manufacturing In Gene Edited Heat-killed Yeast For Insect Control
Read More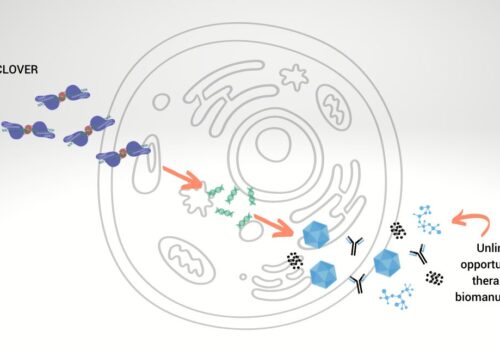
Blog
Cell Bioprocessing
Cas-CLOVER Enables Unlimited Cell Bioprocessing Platform Improvement Opportunities
Read More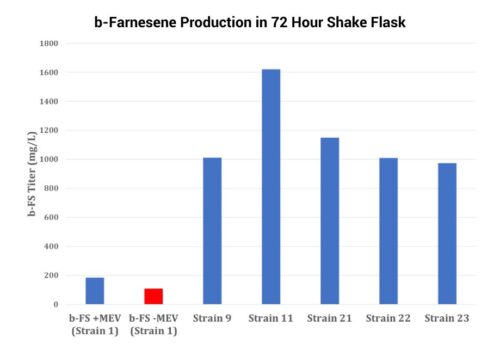
Blog
Synthetic Biotechnology
piggyBac’s Massive DNA Cargo-Carrying Capacity Simplifies And Enhances The Bioproduction Of High Value Metabolites
Read More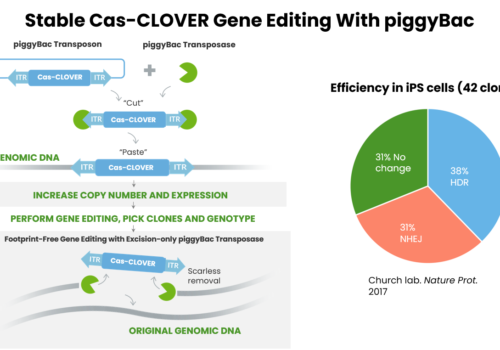
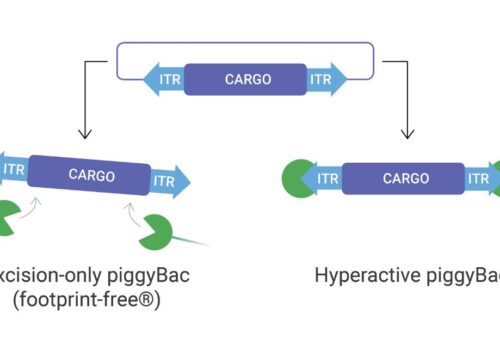
Synthetic Biotechnology
piggyBac: A Simple, Versatile & Efficient Tool For Yeast Strain Development
Read More
Synthetic Biotechnology
Cas-CLOVER: Creating High-Performing Yeast Strains For Industrial Utilization
Read More
Agriculture Biotechnology
Gene Editing in plants is stable, efficient, and reliable with Cas-CLOVER, the clean alternative to CRISPR/Cas9
Read More
Agriculture Biotechnology
Cas-CLOVER: Increasing The Productivity In Crop Trait Engineering The Non-GMO Way
Read More

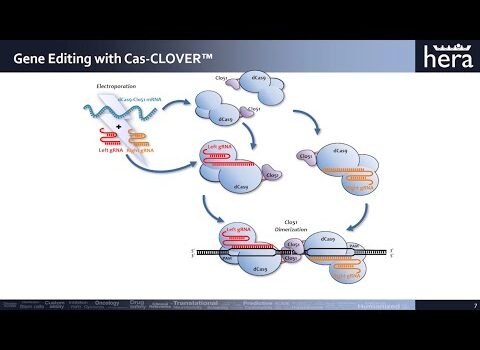
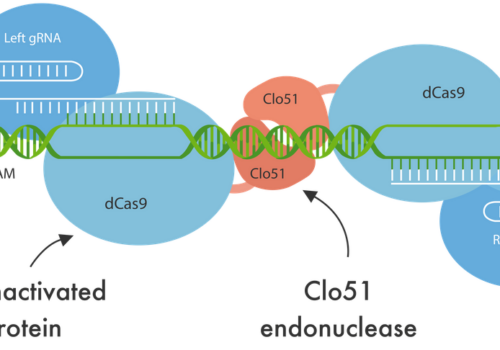
Post Cambridge Webinar: Utilizing Cas-CLOVER piggyBac For Cell Line Development And Engineering Webinar
Read More